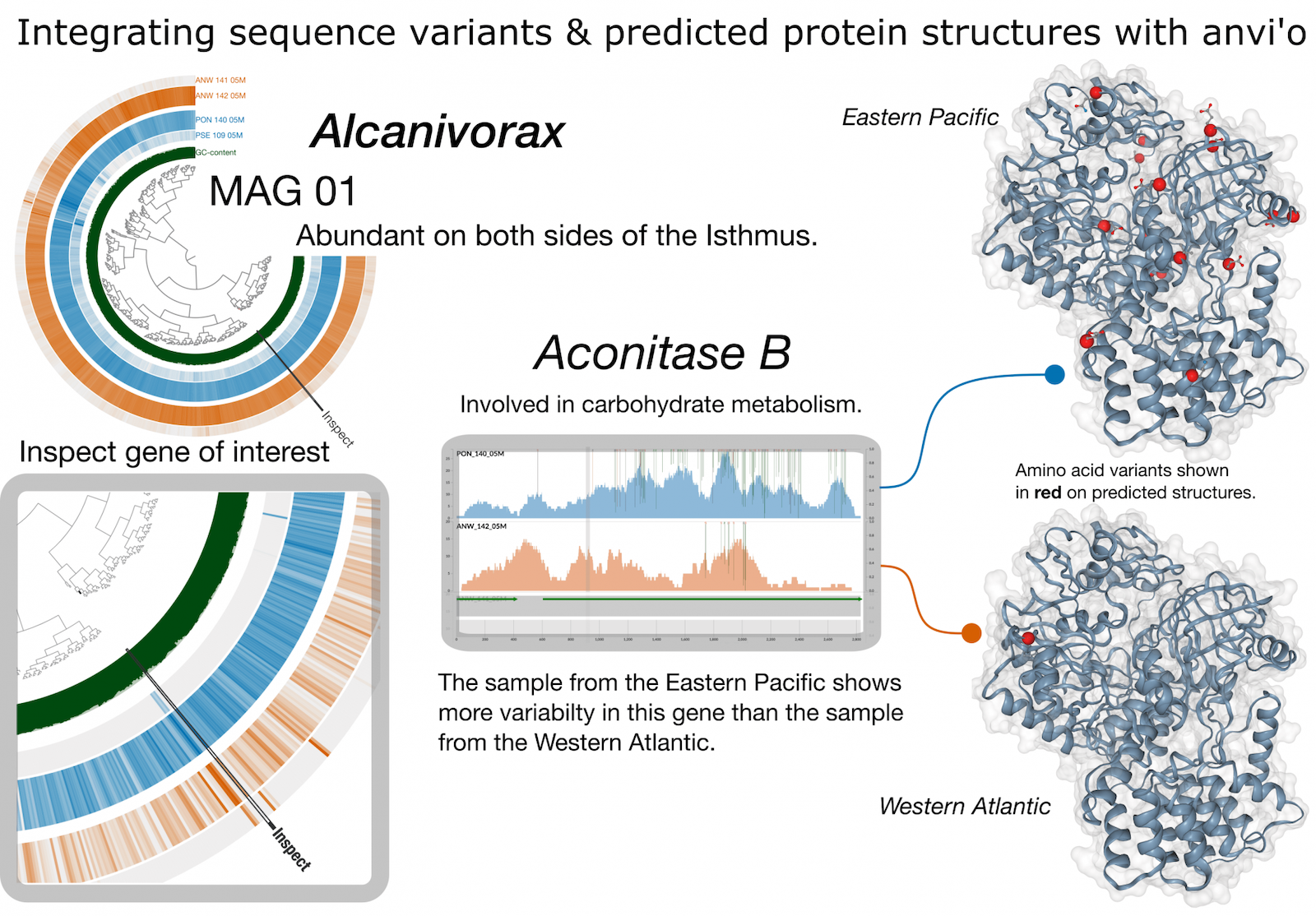
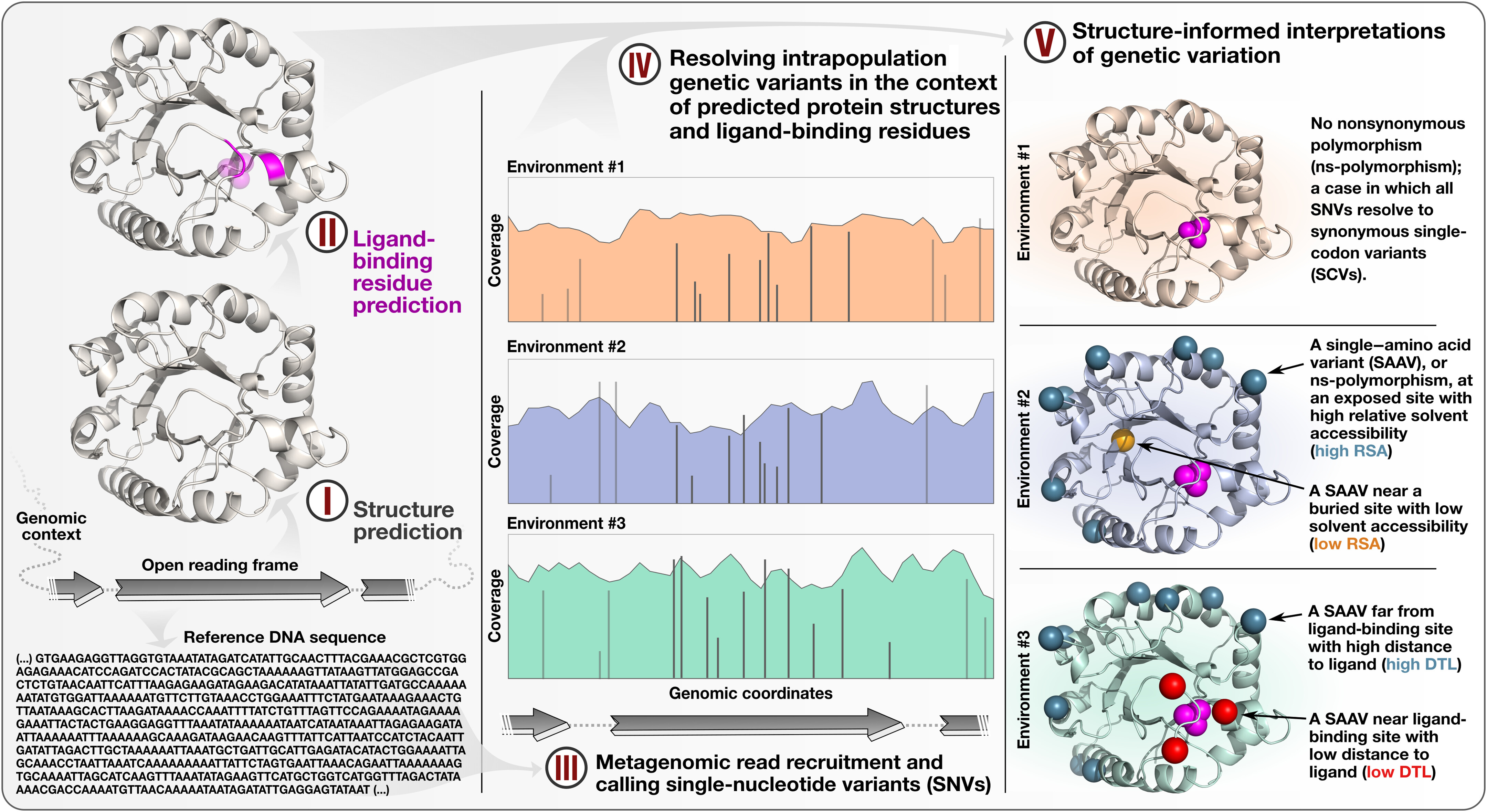
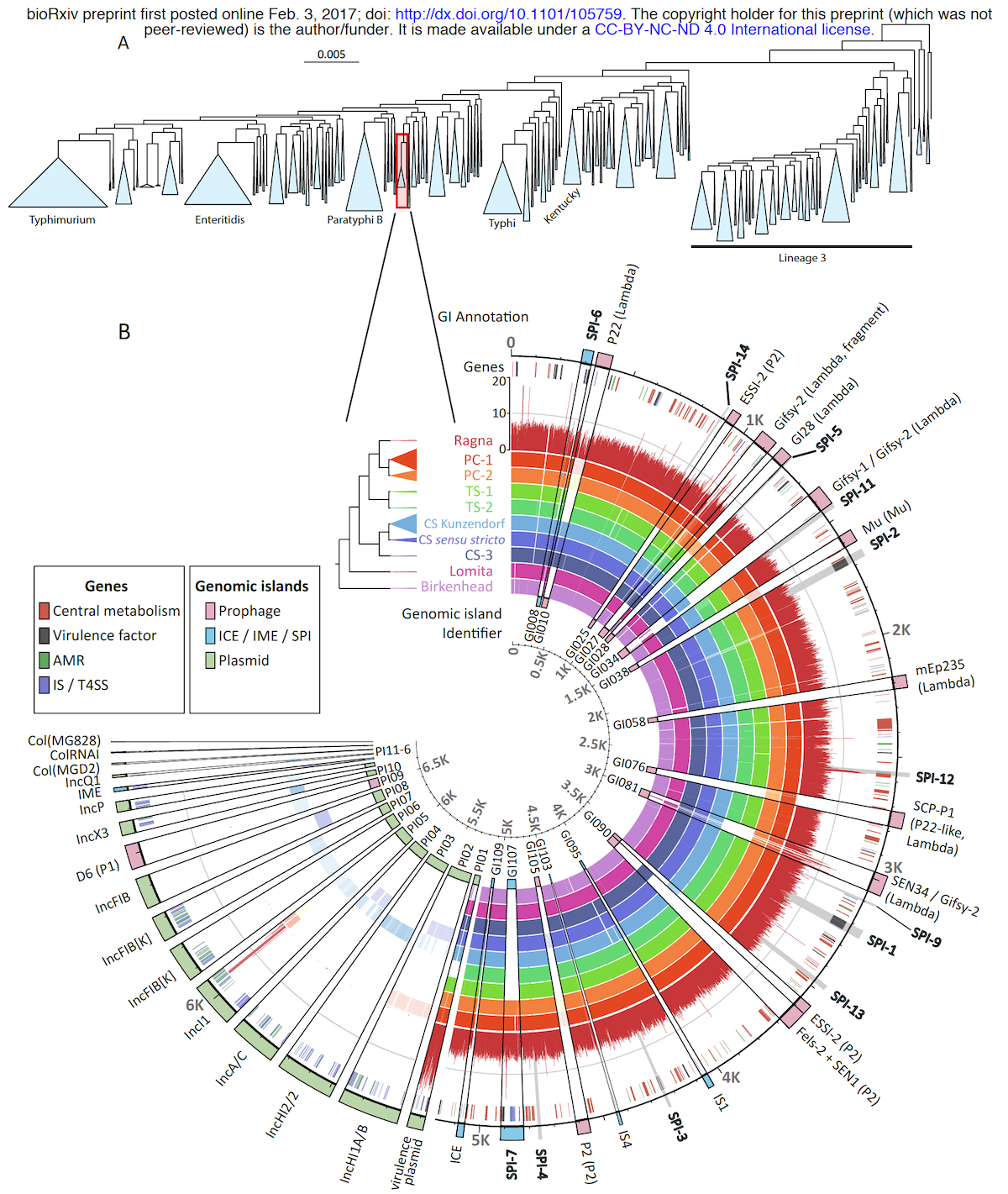
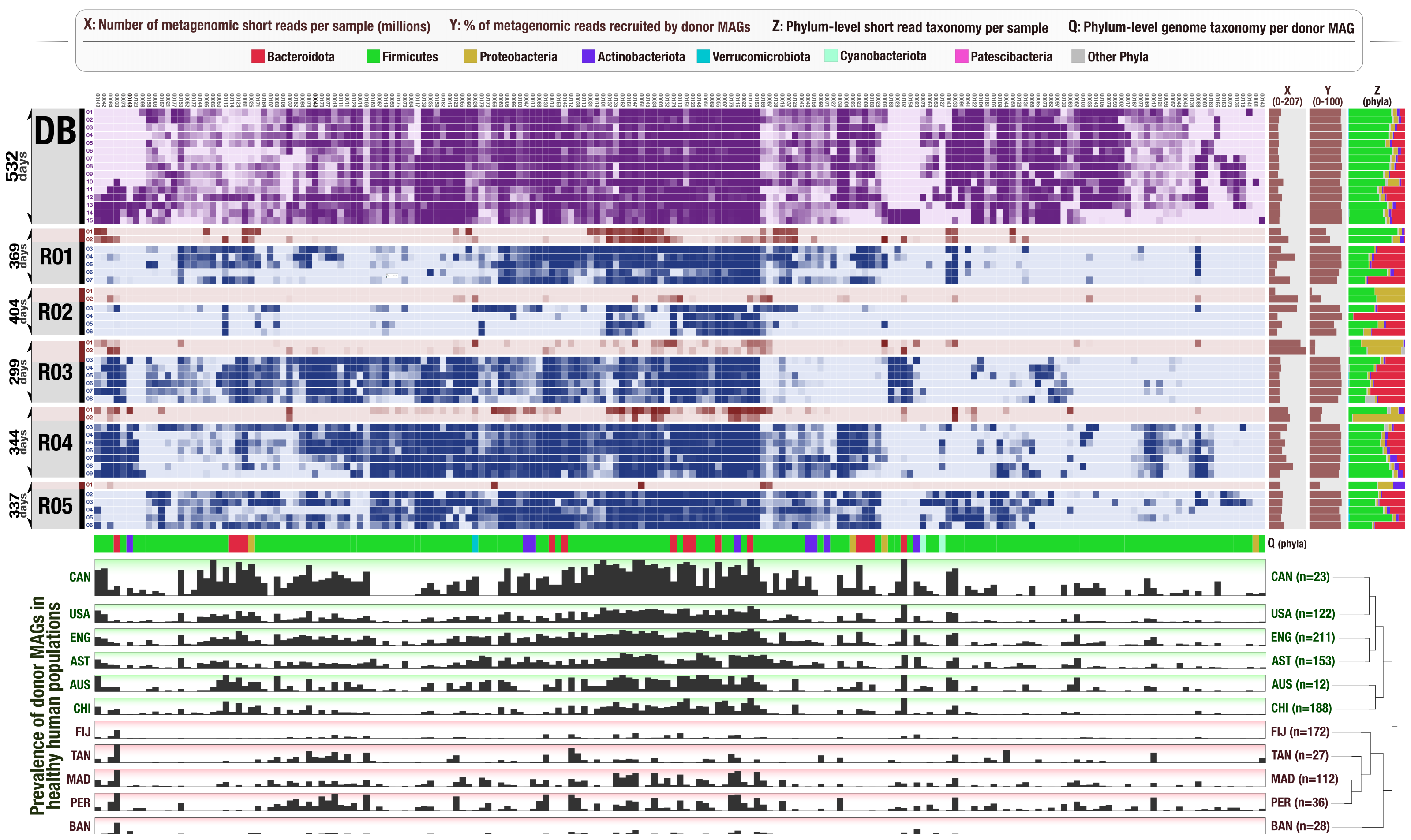
Anvi'o in a nutshell
Anvi’o is a comprehensive platform that brings together many aspects of today’s cutting-edge computational strategies of data-enabled microbiology, including genomics, metagenomics, metatranscriptomics, pangenomics, metapangenomics, phylogenomics, and microbial population genetics in an integrated and easy-to-use fashion through extensive interactive visualization capabilities.
The latest stable version of anvi’o is v9, and you can see the release notes here.
The platform stands on more than 200,000 lines of Python and JavaScript code that follow modern software development paradigms thanks to the voluntary contributions of more than 30 developers from the academia and industry to empower microbiologists for their complex needs through an architecture that enables reproducible science and simple interfaces for data access.
As a principle, anvi’o does not track its users by any means, but based on self-reported data on Twitter, there is at least one anvi’o user in the following countries:
🇺🇸🇨🇦🇬🇧🇦🇺🇨🇳🇳🇿🇸🇪🇨🇭🇵🇹🇯🇵🇮🇹🇫🇷🇪🇸🇩🇰🇩🇪🇨🇱🇧🇷🇦🇷🇺🇾🇹🇷🇪🇬🇸🇬🇸🇦🇷🇺🇳🇱🇵🇷🇵🇭🇳🇴🇲🇾🇲🇽🇲🇫🇱🇺🇰🇷🇮🇳🇮🇱🇮🇪🇭🇰🇫🇮🇨🇼🇨🇷🇨🇴🇦🇹🇨🇿🇦🇪
We welcome you to join in our growing international community, and we look forward to hearing from you. Please share your concerns, ideas, and feature requests with the anvi’o community, or ask your questions on microbial ‘omics on our  group. If you are a programmer, please see this document in the anvi’o repository that covers the platform architecture, major subsystems, coding conventions, and developer workflow.
group. If you are a programmer, please see this document in the anvi’o repository that covers the platform architecture, major subsystems, coding conventions, and developer workflow.
Want to see just a little more? Here is a seminar given by ![]() to introduce anvi’o to the Center for Dark Energy Biosphere Investigations (C-DEBI) by showcasing a few of the anvi’o programs and their applications (if you wish, you can also start from an earlier timepoint to this seminar to have a more comprehensive introduction to the platform).
to introduce anvi’o to the Center for Dark Energy Biosphere Investigations (C-DEBI) by showcasing a few of the anvi’o programs and their applications (if you wish, you can also start from an earlier timepoint to this seminar to have a more comprehensive introduction to the platform).
Funding and Support
Anvi’o is here thanks to the support from,
-
Helmholtz Institute for Functional Marine Biodiversity at Oldenburg (2022-).
-
University of Chicago (2015-2022).
-
Marine Biological Laboratory (2014-2015).
We were able to push the boundaries of anvi’o thanks to the funding we have received from,
-
National Science Foundation (2021-)
via an Integrative Partnership (STC) award for The Center for Chemical Currencies of a Microbial Planet to C-CoMP Team. -
Simons Foundation (2020-2022)
via the Simons Early Career Investigator in Marine Microbial Ecology and Evolution Award to Meren. -
Alfred P. Sloan Foundation (2020-2021)
via the Fellowship in Ocean Sciences to Meren. -
W. M. Keck Foundation (2018-2021)
via the Microbiome dynamics and discovery through transfer RNA to Tao Pan, Meren, et al.
Releases and Codenames
v9 | codename: “eunice”

The code name honors Eunice Newton Foote, an American researcher who discovered the link between atmospheric carbon dioxide concentrations and Earth’s temperature. This groundbreaking discovery emerged from a simple yet powerful experiment: Foote filled one glass cylinder with carbon dioxide and another with ordinary air, then measured their temperatures after exposing them to sunlight. She observed that the cylinder containing carbon dioxide trapped much more heat and remained hot much longer than the control. From this simple observation, she reached an elegant conclusion that established the first connection between carbon dioxide gas and planetary warming. Despite the profound implications of her work, Foote was not able to communicate her findings to the members of the American Association for the Advancement of Science (Joseph Henry presented Foote’s findings to the members of AAAS instead), nor was she recognized as the founder of climate science (that credit went to John Tyndall instead). Her erasure from history was largely due to women’s exclusion from the scientific establishment of her era. We were able to correct the record when retired petroleum geologist Raymond P. Sorenson rediscovered her work after 150 years and sparked renewed recognition of her pioneering role in climate science by publishing an article in 2011 titled “Eunice Foote’s Pioneering Research On CO2 And Climate Warming”. A correction in this instance was only possible thanks to the records of Foote’s work that happened to survive, which is a sobering reminder that for every erased contribution we can recover, many others were likely lost forever. As a generation that experiences the implications of the increasing temperatures her work predicted first-hand, we join others who finally recognize Eunice Newton Foote for her foundational contribution to our understanding of climate change. The code name was a suggestion by Dr. Sofia Ibarraran-Viniegra, managing director of the NSF Science and Technology Center, Chemical Currencies of a Microbial Planet (C-CoMP). See technical release notes.
v8 | codename: “marie”

The code name recognizes Marie Tharp, an American geologist and oceanographic cartographer, who has made immense contributions to earth sciences. Marie was a pioneer in our understanding of oceans as she created the first map of the Atlantic seafloor with her colleague Bruce Heezen. Her work showed that the bottom of our oceans were not only flat sediments but were also covered with canyons, ridges, and mountain ranges that spanned over 65,000 kilometers around the globe. Marie’s revolutionary work emerged from her interpretation of data she was not allowed to collect since women were not allowed to be on ships during the 1950s. Marie compiled her physiographic diagrams from the data Bruce Heezen were able to collect. She did not step on a ship until 1968, and the early evidence she had for seafloor features was initially dismissed as girl talk. The code name was a suggestion by Zena Cardman, a Marine Microbiologist and a NASA Astronaut. See technical release notes.
v7 | codename: “hope”

The code name recognizes Hope E. Hopps as a tribute to all laboratory technicians whose contributions have often been poorly recognized in science. This is despite the fact that technicians not only ensure accuracy, efficiency, and reproducibility in any laboratory, but also push the boundaries of science as much as any other member of their groups, if not more in many cases. Hopps was a specialist in infectious diseases and in 1966 she developed, together with Harry M. Meyer and Paul J. Parkman, a highly effective vaccine for rubella, a viral infection which caused more than 30,000 stillbirths in the United States alone between 1962 and 1965. Despite her role in the vaccine development, in a historical photograph by the NIH that portrays the rubella vaccine development team, Hopps was only identified as “Female Lab Technician“ until recently, even though the caption of the same photograph explicitly named Meyer and Parkman. The unfair treatment of laboratory technicians remains to be commonplace in today’s science. In fact, “not more than a technician’s job” can serve as an argument for professors when they wish to refuse the recognition of one’s contributions to science. We can’t ignore the significant progress we have made as a community during the past few years. But while we continue working on increasing the diversity, equity, and inclusion in science, we must also recognize and face the implicit and explicit biases against those in science who are not PIs, post-docs, or graduate students. The code name was a suggestion by Alon Shaiber, a Genomics Data Scientist at Weill Cornell Medicine. See technical release notes.
v6 | codename: “esther”

The codename is a small tribute to Esther Lederberg (1922-2006), an American microbiologist who studied plasmids and bacterial viruses. Lederberg discovered lambda phage, an E. coli virus that is commonly used in bacterial genetics and molecular biology to deliver DNA into a recipient organism. This led to her description of specialized transduction, that occurs when a prophage improperly excises from the host chromosome carrying host DNA in addition to the viral DNA. In collaboration with her husband, Lederberg developed the technique known as replica plating, which allows repeatable inoculation of bacterial colonies. Lederberg and Luigi L. Cavalli-Sforza discovered the Fertility factor or F-plasmid in E. coli. This is a sequence of DNA that lets the host cell transfer genetic material via a rod-like structure into recipient cells (conjugation). Despite her many incredible scientific accomplishments, she was constantly overshadowed by her husband. She was not appointed to a tenured position while they were both faculty at Stanford, and after their divorce she had a difficult time retaining her appointment. Anvi’o developers dedicate the 6th version of the platform to the memory and revolutionary discoveries of Dr. Lederberg. See technical release notes.
v5 | codename: “margaret”

The codename is a small tribute to Margaret Oakley Dayhoff, an American physical chemist, who is known as the founder of bioinformatics. Dayhoff developed first programmable computer methods to compare protein sequences, and published in 1965 a book titled “Atlas of Protein Sequences and Structure”, which is considered as of today the first text book of bioinformatics. The codename was suggested by Mick Watson, and won the popular vote on Twitter. Dayhoff sadly died at an early age of 57 in 1994, shortly before bioinformatcis emerged as a distinct field. However, her astonishing contributions to life sciences, such as the development of essential approaches for protein sequence comparison and evolutionary tree construction, still constitute some of the most common approaches in our bioinformatics toolkit. See technical release notes.
v4 | codename: “rosalind”

The codename is a small tribute to Rosalind Franklin, the British biophysicist whose work, among other advances in life sciences, contributed to the discovery of the DNA double helix. This codename was inspired by Emily Crossette’s suggestion, ‘esther’, “after Esther Lederberg, who co-developed a replica plating method with her husband but was largely unrecognized and discriminated against as a woman scientist”. Emily explained that her suggestion was to “celebrate how far we have come as a scientific community and look to the future”. Yes. We fortunately did not stay where we were, but we are still far from where we could have been. We remember these women and many others with respect and gratitude, and understand our responsibility to make sure the younger generations of scientists will not suffer from the kinds of discrimination to which their professors were subjected. See technical release notes.