oligotypes
![]()
A CONCEPT-type anvi’o artifact. This artifact is typically generated, used, and/or exported by anvi’o (and not provided by the user)..
🔙 To the main page of anvi’o programs and artifacts.
Provided by
Required or used by
There are no anvi’o tools that use or require this artifact directly, which means it is most likely an end-product for the user.
Description
Oligotyping is a computational strategy that partitions a given set of sequences into homogeneous groups using only a subset of target nucleotide positions.
History
Oligotyping was first described in 2011 and has primarily been applied to 16S ribosomal RNA gene amplicons to resolve closely related but distinct that differ as low as one nucleotide at the amplified region, exceeding the sensitivity of the popular strategy of the time, 97% OTU clustering. Other papers that demonstrate the strengths of this approach include the following: 1, 2, 3, and 4.
The following figure from the oligotyping methods paper depicts major steps of an oligotyping analysis for amplicon sequences:
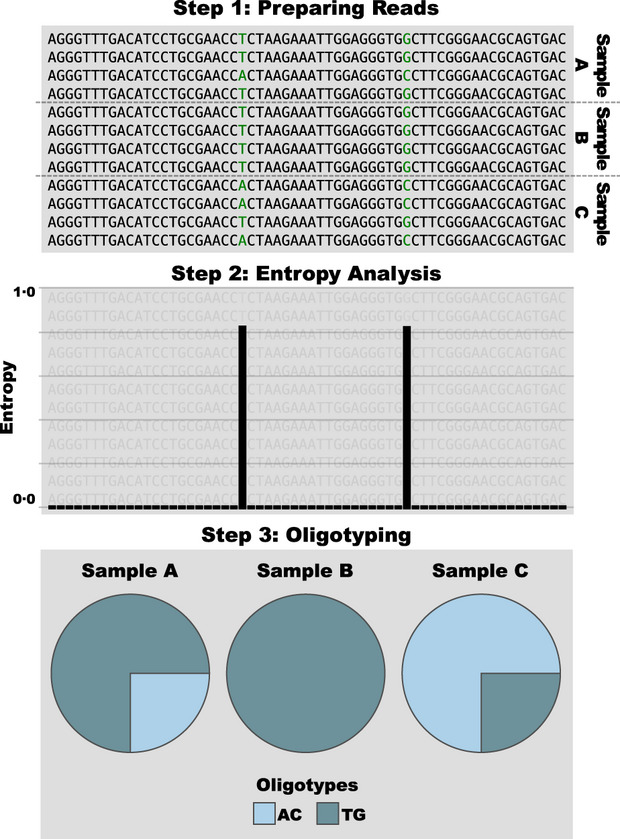
Although, Shannon entropy is not the only approach to identify highly variable nucleotide positions of interest, and they an also be provided by the user.
Applications to metagenomics
This strategy also applies to metagenomic sequences that are mapped to a genomic context to describe the diversity of variable regions that are fully covered by short reads. In the context of metagenomic read recruitment, variable nucleotide positions can be chosen by the user from the positions of single-nucleotide variants anvi’o recovers and presents through inspection pages in the interactive interface or through variability-profile. An example application of oligotyping to metagenomics is demonstrated here:
Edit this file to update this information.