The anvi'o 'contigs' workflow
Table of Contents
From FASTA files to annotated anvi'o contigs databases
This workflow is useful for converting a bunch of genomes into an anvi'o-compatible format. It generates contigs databases from each input FASTA file, and subsequently runs a variety of annotation programs of your choice to populate these databases with some useful information for your downstream work (i.e. functions, single-copy-core genes, taxonomy, etc).
🔙 To the main page of anvi’o programs and artifacts.
Authors
Artifacts accepted
The contigs can typically be initiated with the following artifacts:
Artifacts produced
The contigs typically produce the following anvi’o artifacts:
Third party programs
This is a list of programs that may be used by the contigs workflow depending on the user settings in the workflow-config ![]() :
:
- pyrodigal-gv (Gene calling)
- HMMER (HMM search)
- krakenuniq (Gene taxonomy)
- centrifuge (Gene taxonomy)
- DIAMOND (Sequence search against various databases)
An anvi’o installation that follows the recommendations on the installation page will include all these programs. But please consider your settings, and cite these additional tools from your methods sections.
Workflow description and usage
This workflow is extremely useful if you have one or more fasta files that describe one or more contig sequences for your genomes or assembled metagenomes, and all you want to turn them into contigs-db files.
If you have not yet run anvi’o programs anvi-setup-ncbi-cogs and anvi-setup-scg-taxonomy on your system yet, you will get a cryptic error from this workflow if you run it with the default workflow-config. You can avoid this by first running these two anvi’o programs to setup the necessary databases (which is done only once for every anvi’o installation), or set the rules for COG functions and/or SCG taxonomy to run=false explicitly.
To start things going with this workflow, first ask anvi’o to give you a default workflow-config file for the contigs workflow:
anvi-run-workflow -w contigs \
--get-default-config config-contigs-default.json
This will generate a file in your work directory called config-contigs-default.json. You should investigate its contents, and familiarize youself with it. It should look something like this, but much longer:
and you could examine its content to find out all possible options to tweak. We included a much simpler config file, config-contigs.json, in the mock data package for the sake of demonstrating how the contigs workflow works:
{
"workflow_name": "contigs",
"config_version": "2",
"fasta_txt": "fasta.txt",
"output_dirs": {
"FASTA_DIR": "01_FASTA",
"CONTIGS_DIR": "02_CONTIGS",
"LOGS_DIR": "00_LOGS"
}
}
The only mandatory thing you need to do is to (1) manually create a fasta-txt file to describe the name and location of each FASTA file you wish to work with, and (2) make sure the fasta_txt variable in your workflow-config point to the location of your fasta-txt.
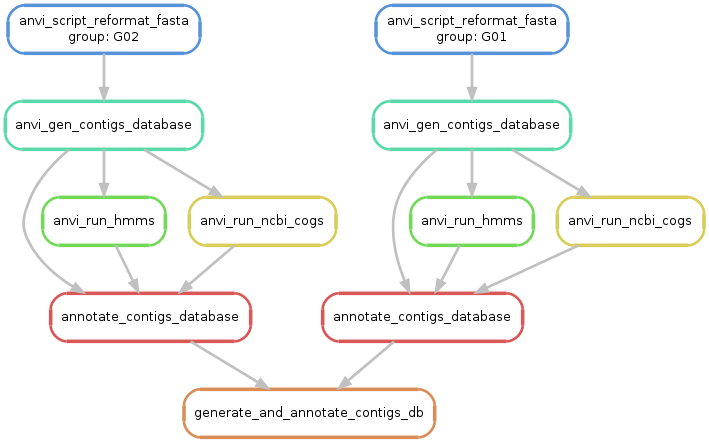
To see if everything looks alright, you can simply run the following command, which should generate a ‘workflow graph’ for you, given your config file parameters and input files:
anvi-run-workflow -w contigs \
-c config-contigs.json \
--save-workflow-graph
For the example config file shown above, this command will generate something similar to this:
Please note that the generation of this workflow graph requires the usage of a program called dot. If you are using MAC OSX, you can use dot by installing graphviz through brew or conda.
If everything looks alright, you can run this workflow the following way:
anvi-run-workflow -w contigs \
-c config-contigs.json
If everything goes smoothly, you should see happy messages flowing on your screen, and at the end of it all you should see your contigs databases are generated and annotated properly. At the end of this process, you will have all your contigs-db files in the 02_CONTIGS directory (as per the instructions in the config file, which you can change). You can use the program anvi-display-contigs-stats on one of them to see if everything makes sense.
Edit this file to update this information.