Some of Us at #ISME18
Table of Contents
The recent ISME Conference at the beautiful city of Lausanne was my very first in-person conference of this scale since the COVID-19 pandemic has started about 42 years ago (or 7 months ago .. really depends on how you perceive and keep track of time nowadays).
My reasons to go to conferences have changed quite dramatically throughout my career in science. I went to the very first science conference in my life during the last year of my PhD. I was told by my mentors at the time that I had to do it because I needed to network. I still have very mixed feelings regarding this term, but I am an adult now and I know when to spare others from my monologues. During the relatively early-middle stages of my career, I went to conferences to share our science and learn about the science of others. While it sounds great in theory, I quickly realized that conferences are one of the least effective means to exchange good science – with so little detail in almost every nicely packaged and well-rehearsed presentation, most talks in mainstream conferences sound a little like science fiction to me. Today, though, as someone who is in the late stages of their career, I find the most powerful aspect of conferences to be the opportunity they present to connect with your colleagues as the people they are, and not just names in papers.
So, I decided to go to the ISME conference this year with that mindset: primarily to connect with some colleagues (while others in our group, Emily Fogarty, Iva Veseli, Sam Miller, and Florian Trigodet did the sharing of our science).
Connecting with colleagues is a natural by-product of every conference, which often happens stochastically. But I decided to try to connect with a particular group of colleagues this time: those who used anvi’o for their research; just so I could learn about them and their research a bit, and ask about their positive and negative experiences with anvi’o to perhaps learn about what anvi’o does well and what needs to be improved. And what could have been a better environment to do that if not finding them in front of their posters. I learned a lot, and everyone received anvi’o stickers as a participation award :p
The purpose of this post is to share some bits from those connections, and introduce some of the members of the anvi’o user community to you, along with their research and their take on anvi’o.
I hope you will find this post beneficial to recognize some new names and faces and their research, and/or to hear some positive and negative testimonials on anvi’o.
Unfortunately I couldn’t make it to every single poster I wanted to visit during ISME. I am sorry for that. But I hope there will be other opportunities in the future.
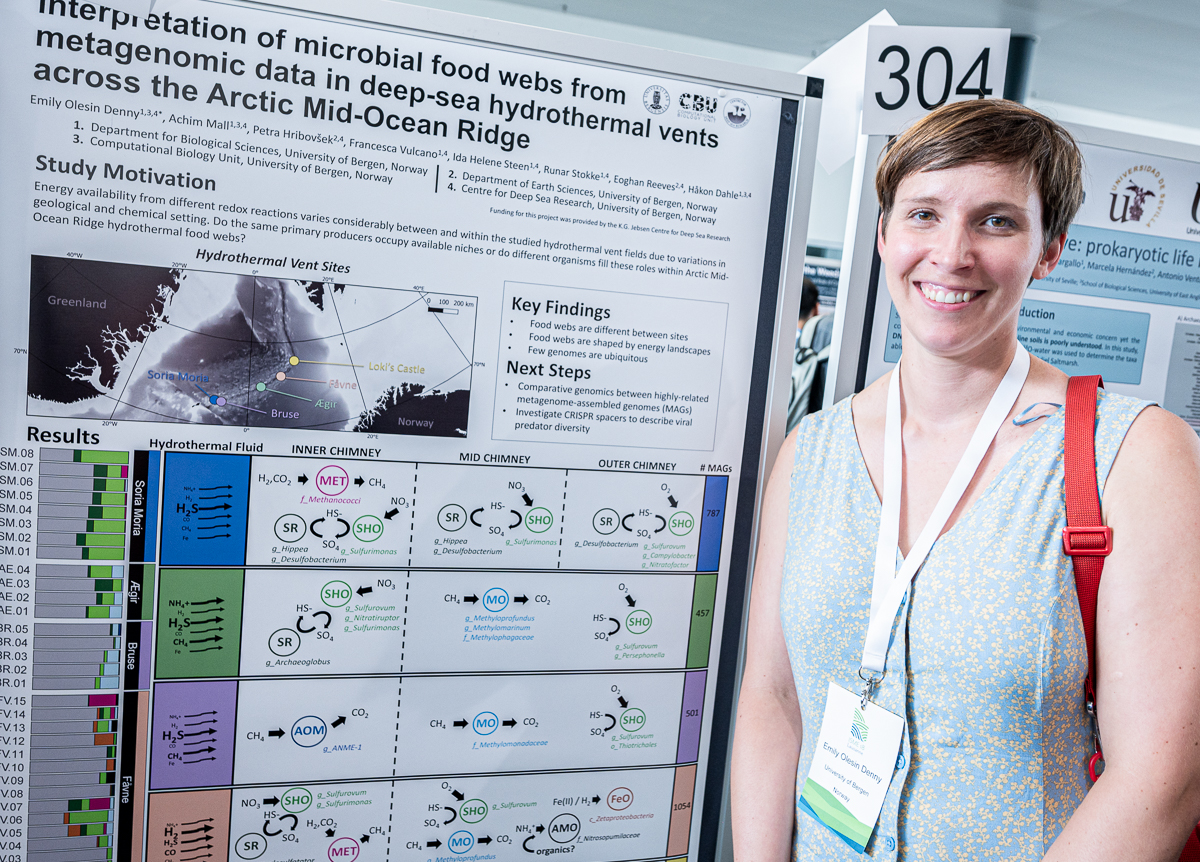
Emily Olesin Denny
Emily’s poster at #ISME18 titled ‘Interpretation of microbial food webs from metagenomic data in deep-sea hydrothermal vents across the Arctic Mid-Ocean Ridge’. When I asked what was the most interesting finding in this poster, Emily mentioned that despite many similar microbial niches that exist across different deep-sea hydrothermal vent sites, they found only a few genomes that were shared across all of them.
Emily Olesin Denny (@EmmaLeeDenny) is a computational biologist, and currently a PhD candidate in the Dahle Group at the Computational Biology Unit of the University of Bergen, Norway. Emily is broadly interested in understanding which biological signatures have the tightest relationship with an ecosystem’s energetic constraints, and why.
Emily has been an anvi’o user for less than 2 years, and has been using the platform primarily for processing and visualizing metagenomes. I asked Emily “what do you like most about anvi’o (if you can think of anything you like about it)?” and got this response:
I like that anvi’o tools are so portable and come with such detailed and cheeky documentation!
I also asked Emily “what would you say about anvi’o to someone who never used it before to warn them?”. Just to learn more about some of the shortcomings of the platform. Emily responded,
I think I’d tell someone who is doing large metagenomics projects like mine that co-mapping is probably the way to go rather than co-assembly, but this methodology is not well documented among the anvi’o user community. Just a few Slack posts here and there. As the data gets cheaper to produce more modular methods of analysis will be needed.
I absolutely agree with this sentiment. While pretty much anything is possible with anvi’o, unfortunately our documentation is not quite up-to-date with all possible ways to use anvi’o for genome-resolved metagenomics. We often use single assemblies ourselves, and I hope we will be able to improve our documentation further soon.
It turns out, if it was absolutely necessary to choose a different career path outside of research, Emily would have chosen to to be an illustrator :) I also asked Emily “given all the time you have spent in science, what advice would you have given to your younger self?”. No wiser words have ever been spoken:
Don’t do it perfectly. Just do your best, then keep rolling.
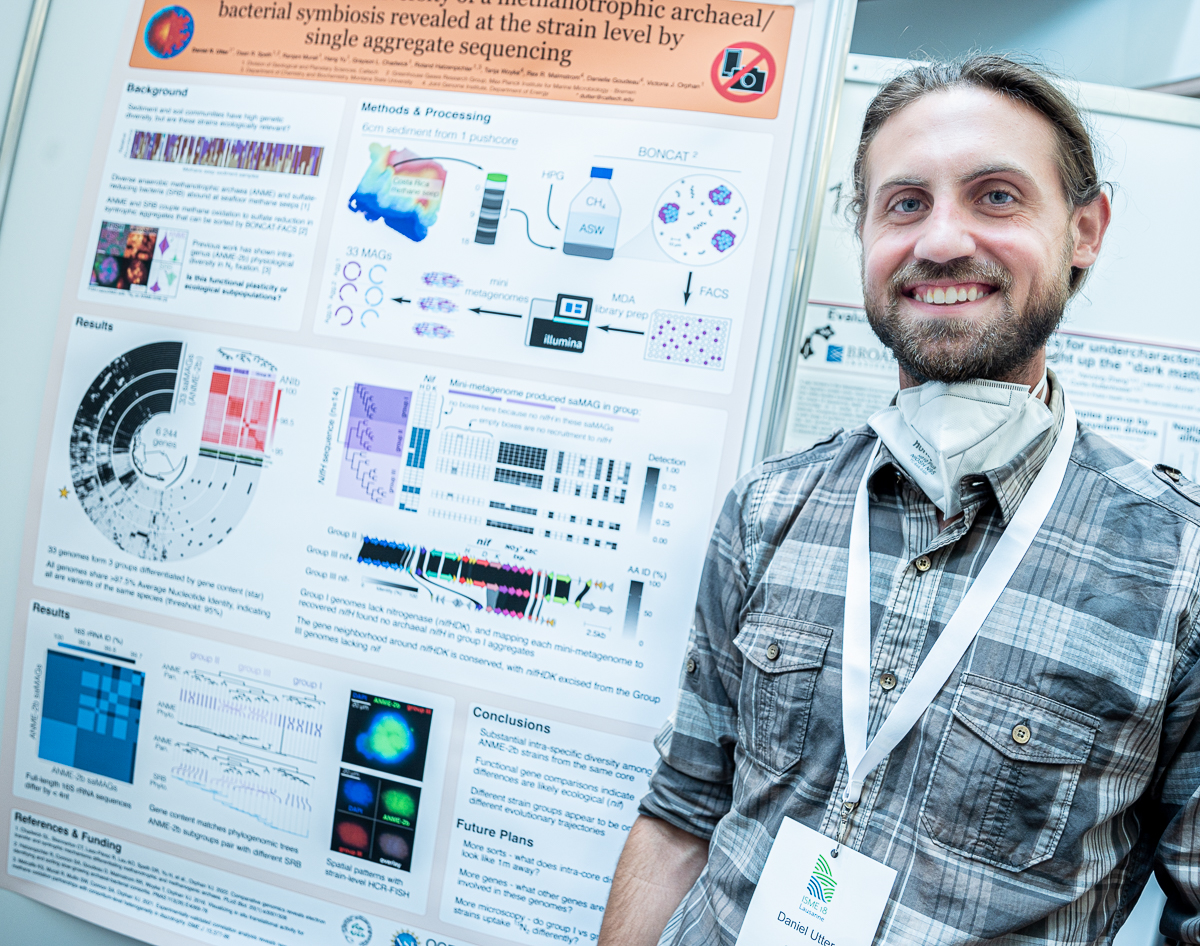
Daniel R. Utter
Dan’s poster at #ISME18 titled ‘Genomic and functional diversity of a methanotrophic archaeal/bacterial symbiosis revealed at the strain level by single aggregate sequencing’. When I asked what was the most interesting finding in this poster, Dan said ‘just six centimeters of mud holds 3 subspecies of a single species, each of which with distinct genomic adaptations for different ecological strategies based on nitrogen’.
In Dan’s poster you clearly can see the “no photographs, please” sign. I know it is there for a reason, but please trust me that I did ask the permission of each presenter to take and share their photographs :) Dan kindly mentioned that I could remove it from the poster digitally if I wanted, and I am very sorry for deciding to not do that and instead waste your time to read this disclaimer :)
Daniel R. Utter (@bajenak) is a microbial ecologist and, in Dan’s own words, a bioinformatician wannabe – although those of us who are familiar with Dan’s work and commitment to open and reproducible bioinformatics know that it is just too modest of Dan to say that.
Dan is currently a postdoctoral scientist at the Orphan Lab at the California Institute of Technology, United States, and generally interested in understanding the ecological forces shaping the evolution of in situ microbial populations in different systems, and how does the relevance of these factors change from micro to macro scales.
Dan has been an anvi’o user for more than 2 years, and has been using the platform practically for everything ‘omics: metagenomics, reconstructing genomes from metagenomes, curation of existing genomes, generating reference pangenomes, metapangenomes, studying population genetics through single-nucleotide variants, single-codon variants, and single-amino acid variants. Dan also run a few workshops for general bioinformatics/metagenomics workshops in which anvi’o was a backbone for ‘omics. Essentially one of the more advanced users of the platform. I asked Dan “what do you like most about anvi’o?” and got this response:
My main use is the management of all data stored in anvi’o (contigs, gene calls, functions, sequence coverage, etc.) in relation to one another. This integrated access to data has saved me so many times from indexing something wrong in Python…
The ease of switching scales in anvi’o visualizations is also really nice – being able to go from contig-scale coverage to nucleotide-scale coverage, BLAST searching genes with a single click etc, are all super nice.
The functional enrichment feature is incredibly handy. I also appreciate how the output is not only statistical or only descriptive regarding which genomes have which functions, but both, so I can use the same output in multiple ways depending on my question.
Managing anvi’o on shared computer systems is also quite straightforward.
It is a fact that those of us who are able to solve their own problems with quick-and-dirty scripts tend to avoid software platforms designed for end-users. Since software solutions designed to be easy-to-use can limit power users. But I think the points Dan makes about anvi’o are great examples of how power users too can benefit from anvi’o. But of course anvi’o is far from being perfect:
The abundance of reserved language is necessary, but unfortunately makes the learning curve noticeable. E.g., the allocation of information to an anvi’o contigs-db versus a profile-db is natural once you know what’s going on, but as a new user (or a teacher trying to explain to a new user) understanding why they are separate and what information is where can be a bit of a pain to figure out. And element names such as ‘items’ or ‘layers’ make sense once you get used to the anvi’o interactive interface, but they can be confusing at first.
It’s totally worth it and seems necessary, and the improvements in key-wording on the online anvi’o documents are major helps, but to me, figuring out the internal language was the biggest impediment (disclosure: having an internal language is 100% necessary because the field doesn’t have consistent, metagenomics-sensible definitions for terms such as population, gene, etc. Just musing on my UX thoughts).
This sentiment resonates with me quite a bit. For instance, we called the contigs database contigs database, because it was an anvi’o data product that contained one or more contigs. But more recently in the C-CoMP (which is the new NSF Science and Technology Center for Chemical Currencies of a Microbial Planet that you certainly need to come join if you are interested in such stuff) we faced some terminology challenges. More specifically, we have been exploring the use of anvi’o contigs databases to share information about our model organisms between different ends of the science spectrum to make sure all participants of this Center effort would be using the same data. But the unfamiliar terminology certainly adds an additional barrier bring together biochemists, mathematical modelers, microbial ecologists, microbiologists, and more. For instance, when we tell someone “here is a contigs database for Ruegeria pomeroyi”, the first question is often “what is a contigs database”. This is a difficult problem to solve since what makes sense from a computational and/or software design perspective may be very counter-intuitive or meaningless to life scientists and others. We are open to suggestions regarding how to improve our documentation and language.
Going back to Dan, I also learned that if Dan had to choose a different career path outside of research, it would’ve been orchid farming and conservation :) How refreshing.
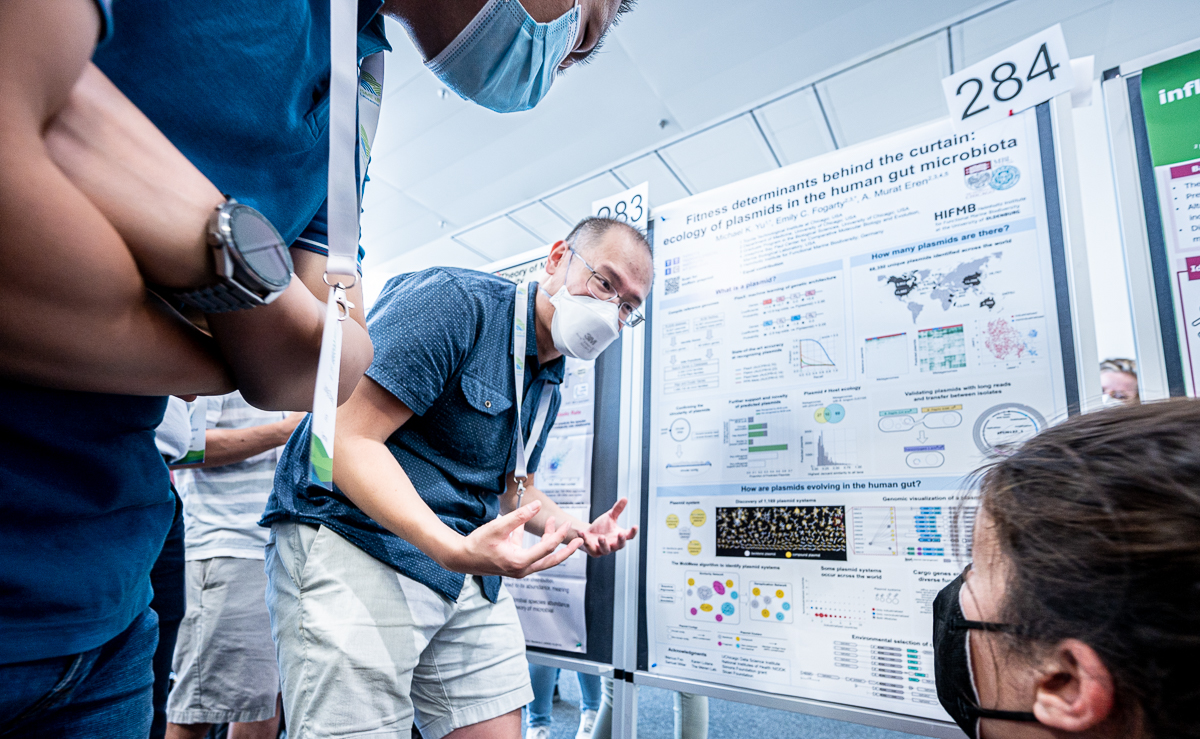
Michael K. Yu
Mike Yu’s poster at #ISME18 titled ‘Fitness determinants behind the curtain: ecology of plasmids in the human gut microbiota’. When I asked what was the most interesting finding in this poster, Mike Yu said ‘The human gut is home to 200 times as many plasmids as known plasmids, and we now have the computational methods to study the diversity and ecology of plasmids in any microbial environment.’
Michael K. Yu (@michaelkuyu), a long-term collaborator and a friend of our group is a computational and systems biologist, and currently a Research Assistant Professor at the Toyota Technological Institute at Chicago. Mike is generally interested in developing machine learning and bioinformatics methods to study the diversity of life. Even though Mike is trained as a computer scientist, he is deeply interested in understanding life rather than cutting corners in computation.
Mike has been an anvi’o user for more than 2 years, and has been using the platform primarily to study metagenomic assemblies, gene functions, and to profile metagenomic read recruitment results. I asked Mike “what do you like most about anvi’o (if you can think of anything you like about it)?”:
The anvi’o workflows make it very easy to run and reproduce a series of established bioinformatics analyses of metagenomes.
But there is always room for improvement, especially for power users:
The anvi’o scripts are really helpful and well documented, but I often want to run them directly in a Python kernel (e.g. in a Jupyter notebook) rather than create a new terminal subprocess.
It would be nice if every terminal command for running an anvi’o script had an equivalent command in Python.
A nice challenge for anvi’o developers (these suggestions often made me think about creating a list of small projects and describe them in great detail on a place like GitHub for those who wish to join the effort to have some ideas about what they could be doing).
If it was absolutely necessary to choose a different career path outside of research, Mike would have chosen to develop indie video games, and start a restaurant that specializes in many types of asian noodle soups :) I also asked Mike “given all the time you have spent in science, what advice would you have given to your younger self?”:
(1) If you plan to eventually become a professor and stay in academia, then finish your PhD as fast as possible with the bare minimum requirements. And then, concentrate on publishing a lot in your postdoctoral/post-PhD position (and spend more years in this position rather than the PhD). Sadly, professors on hiring committees seems to judge you mainly on your output during your postdoc rather than your PhD (no matter how much you produced in your PhD).
(2) Burn out is real. Make sure you have regular hobbies that enrich your life beyond your immediate work. Life is beautiful – too beautiful for work to get in the way of appreciating it.
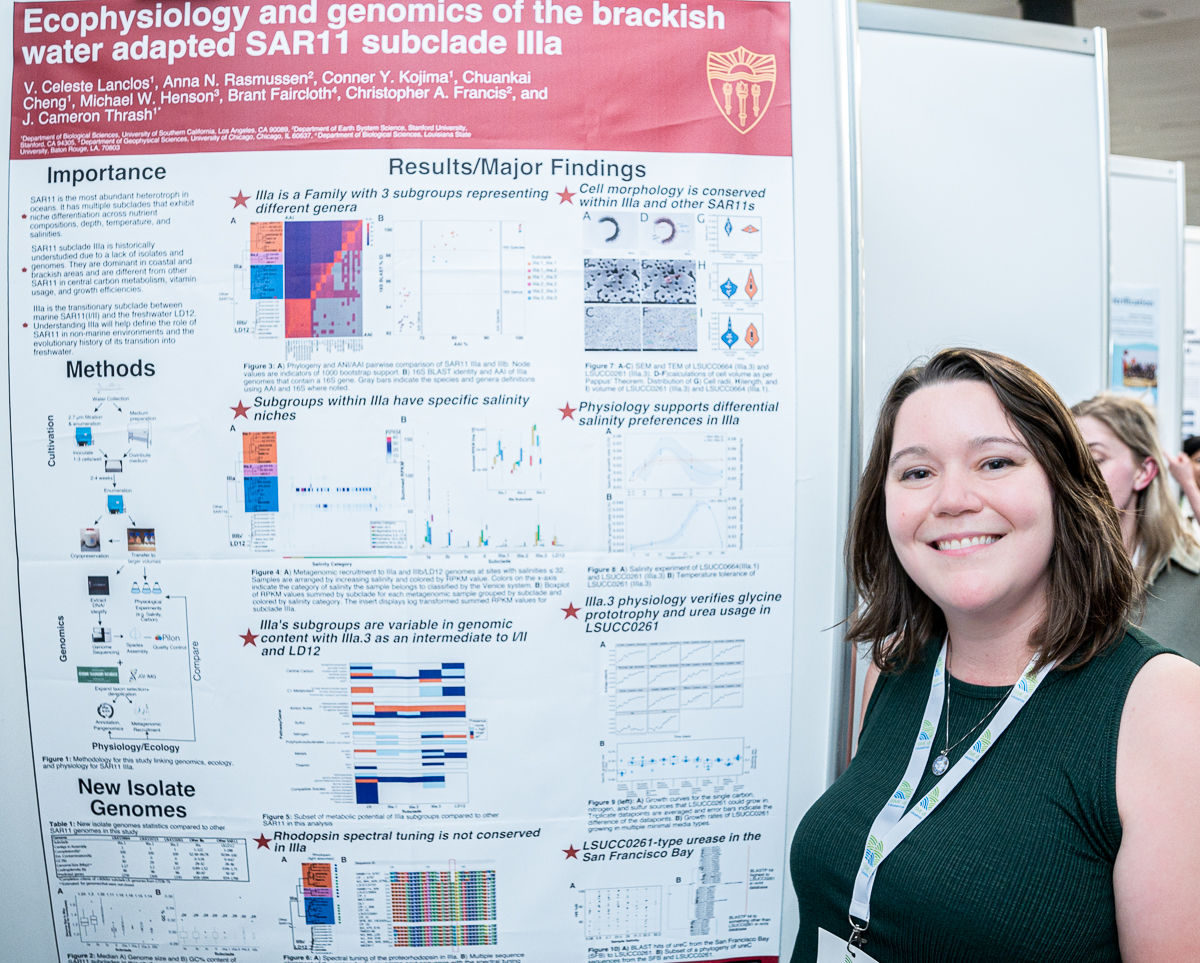
V. Celeste Lanclos
Celeste’s poster at #ISME18 titled ‘Ecophysiology and Genomics of the Brackish Water Adapted SAR11 Subclade IIIa’. When I asked what was the most interesting finding in this poster, Celeste said ‘we were finally able to confirm that the salinity specialization of IIIa seen in ecological data is reflected in pure cultures; the two cultivated subgroups have differing salinity ranges and also differ in growth rates for overlapping salinities.’
V. Celeste Lanclos (@vclanclos) is a microbial ecologist, and currently a PhD candidate at Cameron Thrash’s Group at the University of Southern California and generally interested in understanding the interplay between cultivar-informed genomics/ecology and the genomics/ecology-led cultivation of understudied environmental microbes.
Celeste has been an anvi’o user for more than 2 years, and has been using the platform primarily for pangenomic analyses of diverse clades of bacteria. So I asked Celeste “what do you like most about anvi’o (if you can think of anything you like about it)?” and got this response:
Anvi’o really fills a niche that we needed to revolutionize comparative genomics. What we previously did for this kind of work was clunky with a ton of EXCEL sheets and manual curation of the contents of gene clusters. Anvi’o really has made my life more pleasant.
I also love when the error messages get a little sassy =)
Anvi’o developers would like to apologize for the general attitude of the platform. Celeste had some fair warnings regarding the limits of the visaulization of pangenomes in anvi’o:
I crashed the visualization portion of the workflow many times with my 471 genomes. When I had fewer genomes, the interactive interface was a little hard for me to understand how to best use.
It is true that the anvi’o interactive interface can be challenging to use for large-scale analyses of pangenomes. It is more or less a design decision where we aim for precision and detailed analyses of subtle patterns through interactive interfaces rather than summaries of large-scale data. That’s why even though anvi’o can still compute pangenomes from hundreds of genomes and enable non-interactive analyses of pangenomes, the interactive interface will disappoint you increasingly with increasing numbers of genomes :(
If it was absolutely necessary to choose a different career path outside of research, Celeste would have chosen to be a writer :) I also asked Celeste “given all the time you have spent in science, what advice would you have given to your younger self?”, and heard some wise words:
The gap between where you are and where your peers/supervisors are is not all that big after all. With time and humility, you’ll get there too.
Hyper-focusing on the distance between where you are and where you want to be only leads to stagnation.
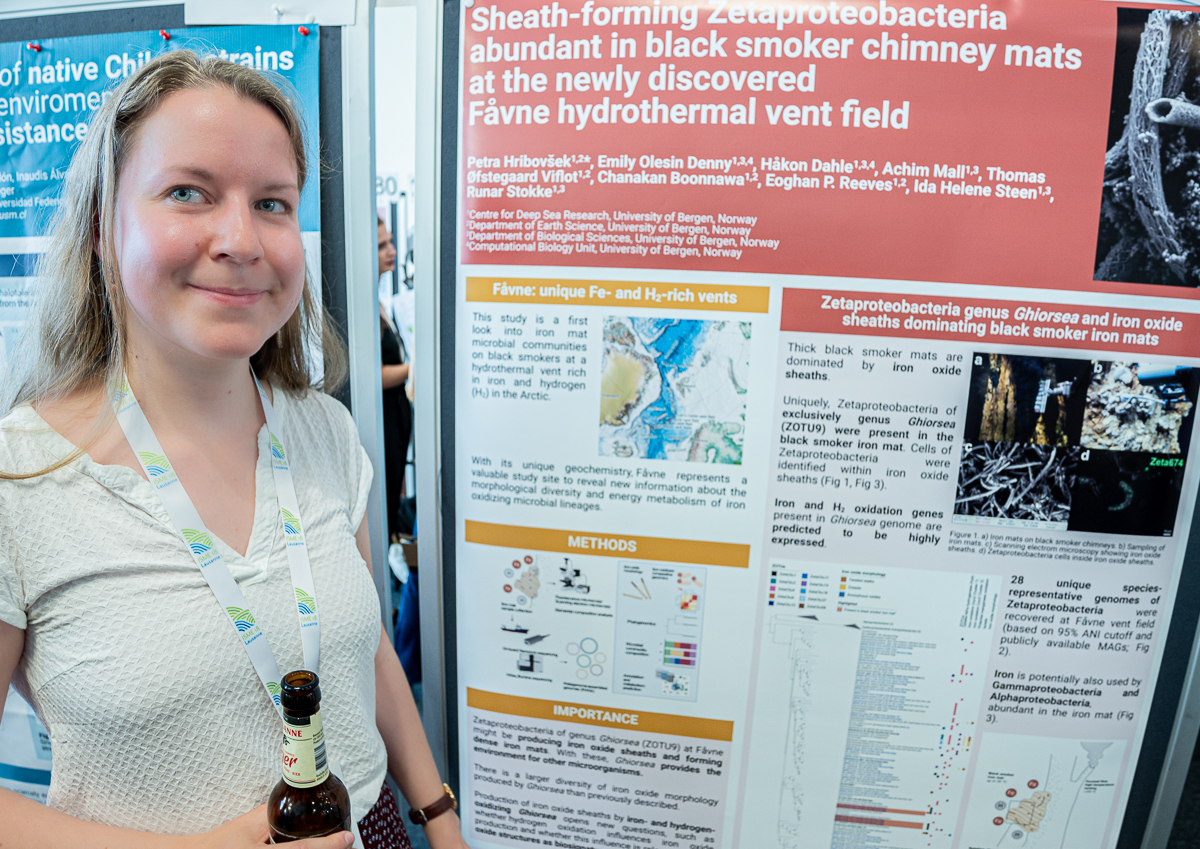
Petra Hribovšek
Petra’s poster at #ISME18 titled ‘Sheath-forming Zetaproteobacteria abundant in black smoker chimney mats at the newly discovered Fåvne hydrothermal vent field’. Regarding the most interesting finding in this poster, Petra said ‘we now have an idea about which bacteria are producing iron oxide tubular structures that form dense microbial mats at hydrothermal vents.’
Petra Hribovšek (@petrahribovsek) is a microbial ecologist, and currently a PhD candidate at Centre for Deep Sea Research at University of Bergen, and generally interested in understanding how microbes affect their environment.
Petra has been an anvi’o user for less than 2 years, and has been using the platform primarily for manual refinement of metagenome-assembled genomes, comparative genomics, pangenomics, phylogenomics, and for quick visualization needs. I asked Petra “what do you like most about anvi’o (if you can think of anything you like about it)?” and got this response:
Seeing genomes being visualized with anvi’o the first time helped me to grasp the concepts within genomics and metagenomics (manual binning of genomes, pangenome visualization).
I like anvi’o since everything is in one place and one can easily do quite many analyses. Anvi’o is also constantly evolving and adding new functions. Among several things, I appreciate the possibility to import your own annotations and gene calls, function enrichment analyses, and the ease of recovering single-copy gene sequences for phylogenomics.
And last but not least, anvi’o has really helpful and kind developers and a community to contact when stuck.
I also asked Petra “what would you say about anvi’o to someone who never used it before to warn them?”
Since I want to use different ways to analyse genomes, it took time to adapt everything to anvi’o.
Although we do our best to make sure it is easy to enter into the anvi’o ecosystem with different kinds of data (from user-defined gene calls to functions), there is a lot of room for improvement there to be able to quickly integrate information stored in file formats such as GFF3, GenBank, etc. We are always keen on hearing about such struggles to see if we can improve things.
If it was absolutely necessary to choose a different career path outside of research, Petra would have chosen to walk on a path to work towards ensuring environmental sustainability, helping people, and managing projects :) I also asked Petra “given all the time you have spent in science, what advice would you have given to your younger self?”:
Failing is good. Ask questions, dive into science, find your community. And be kind to yourself.
Haris Zafeiropoulos
Haris’s poster at #ISME18 titled ‘Reverse ecology and systems biology approaches to identify key processes ensuring life in extreme environments’. When I asked what was the most interesting finding in this poster, Haris said ‘I would say the role of anaplerotic reactions in a hypersaline mat that we were studying: how seasonality is linked to the functioning of microbial communities fascinates me’
Haris Zafeiropoulos (@haris_zaf) is a bioinformatician, who just defended a PhD at Evangelos Pafilis’s Group at the Hellenic Centre for Marine Research, and about to start a postdoctoral appointment with the Microbial Systems Biology Lab led by Karoline Faust at the Rega Institute, Belgium. Haris is generally interested in inferring microbial interactions in dynamic microbial systems to understand the determinants of community fitness, and whether evolution can play a part in this at higher levels of taxonomy.
Haris is new to anvi’o and has been a user of the platform for less than 6 months, primarily using it for the refinement of metagenome-assembled genomes and functional annotation. I asked Haris “what do you like most about anvi’o?” and got this response:
More than anything, I enjoy the anvi’o community. As a user I felt that there is always someone there to guide me and propose alternatives, and as a developer, even from the outside, it feels that it is a great environment that represents open-source at its best.
Regarding its modularity, I think I really liked the database-oriented perspective of anvi’o that allows you to do so many different things at the same time. I also liked the way KEGG modules are handled and reported. And of course, the approach that makes the user responsible for what is going on.
It is refreshing to hear that the strictly hands-off approach of anvi’o where the platform avoids making any decisions on behalf of the user behind the scenes, but forces the user to be on top of their analyses. Which can initially be a difficulty to overcome, but we all believe it is for the best to let the user to be solely responsible for their analyses.
I also asked Haris “what would you say about anvi’o to someone who never used it before to warn them?”
It was quite easy for me to start working with anvi’o especially since I watched a few videos that I believe were quite important to me to realise its philosophy.
I would strongly suggest people to first find an HPC or a server to work with and not try anvi’o on personal computers.
I think it is certainly essential to have an HPC/server solution accessible for large-scale analyses. We often generate anvi’o data products (such as contigs databases and profile databases) on servers, and then download those data products to our personal computers to work with them interactively.
If it was absolutely necessary to choose a different career path outside of research, Haris would have chosen to sell books in outdoor markets, and drive a truck in the Balkans :) I also asked Haris “given all the time you have spent in science, what advice would you have given to your younger self?”:
Study more before you start implementing things, and always make arrangements with the group you are working with about the crucial parts of a study.
Don’t take high-impact factor journals too seriously as you will find good research almost everywhere as science publishing is quite an industry.
There is no reason to do that project if you don’t like it.
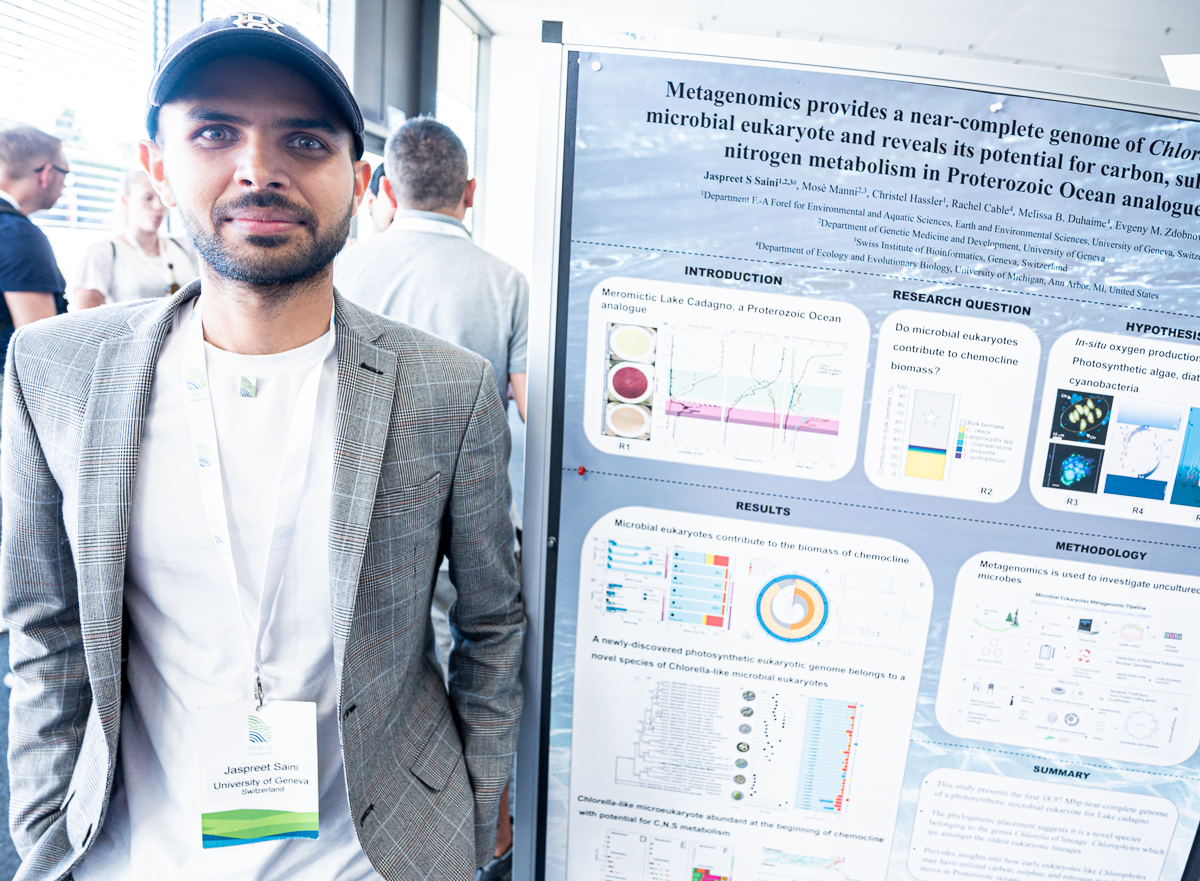
Jaspreet Singh Saini
Jaspreet’s poster at #ISME18 titled ‘Metagenomics provides a near-complete genome of Chlorella-like microbial eukaryote and reveals its potential for carbon, sulphur and nitrogen metabolism in Proterozoic ocean analogue’. When I asked what was the most interesting finding in this poster, Jaspreet said ‘Microbial eukaryotes can have versatile C, N, and S metabolism. And they can also contain phycobilisomes, often presumed to be cyanobacterial specific.’
Jaspreet Singh Saini (@jaspreet0710) is a microbial ecologist. Until recently Jaspreet has been a part of the Duhaime Lab of Aquatic Microbial Ecology at the University of Michigan, United States, and he is now a postdoctoral scientist with Christof Holliger’s Group at the EPFL, Switzerland.
Jaspreet is generally interested in the early evolution of life on our planet and has been using anvi’o for more than 2 years for all sorts of ‘omics. This is what Jaspreet liked most about anvi’o:
Power of control for each step of metagenomics, and the ability to manually refine metagenome-assembled genomes through interactive visualizations.
I also asked Jaspreet “what would you say about anvi’o to someone who never used it before to warn them?”
It is not easy to assign taxonomy to microbial eukaryotes in anvi’o.
Anvi’o can indeed do much better with eukaryotic organisms. Something we are looking forward to addressing it ASAP not only because many of our colleagues need it from anvi’o, but also larger efforts such as C-CoMP demands better integration across major clades of life.
If it was absolutely necessary to choose a different career path outside of research, Jaspreet would have chosen to be a classic chef :) I also asked Jaspreet “given all the time you have spent in science, what advice would you have given to your younger self?”. Here is the response:
Take time for yourself and your well-being. Go out with your family, friends, and relatives. Pick any sport for your physical health or do yoga. Let science wait sometimes.
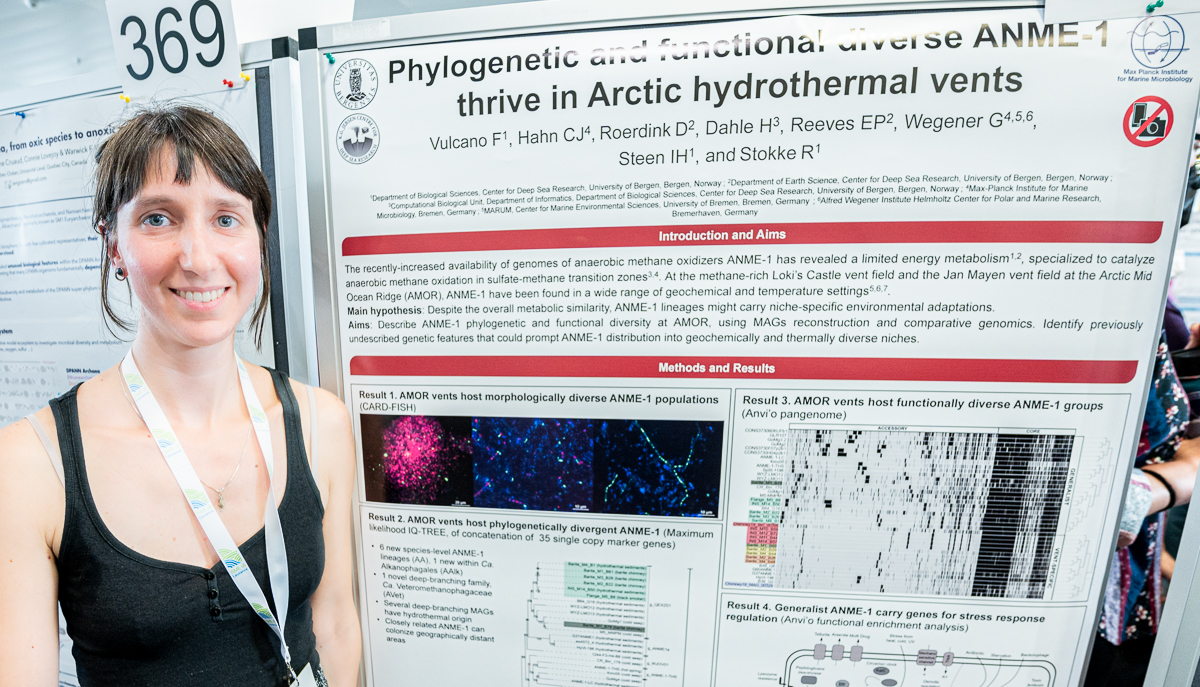
Francesca Vulcano
Francesca’s poster at #ISME18 titled ‘Phylogenetic and functional diverse ANME 1 thrive in Arctic hydrothermal vents’. When I asked what was the most interesting finding in this poster, Francesca said ‘genetic systems involved in regulating metabolic activity might favor dispersal of anoxic marine microbes’
Francesca Vulcano (@FrancescaVulca2) is a molecular biologist and a microbiologist, and currently a PhD candidate at the Centre for Deep Sea Research of the University of Bergen and interested in understanding the evolution of metabolic pathways the role and ratio of the ‘chance’ versus ‘necessity’ behind their evolutionary trajectories.
Francesca has been an anvi’o user for less than 2 years, but has already been using it for many things, including annotating genomes, extracting markers for phylogenomics, pangenomics, and comparative genomics through functional enrichment analyses. I asked Francesca “*what do you like most about anvi’o”:
I like anvi’o because it is extremely user-friendly. I have no bioinformatics background or any particular skills in informatics. Nevertheless, anvi’o makes it possible for me to run analyses that are now crucial and basic requirements in the field of microbial ecology. The tutorials are detailed and the community of developers is easy to reach.
I also asked Francesca “what do you think we need to improve in anvi’o?”, and I was met with resistance :)
I honestly don’t see any issue with anvi’o. Of course, the more bioinformatics skills you have, the more you get out of the platform, but that’s mostly up to the user :) Anvi’o has been great for my purposes so far.
If it was absolutely necessary to choose a different career path outside of research, Francesca would have chosen to be an illustrator and comic writer :) I also asked Francesca “given all the time you have spent in science, what advice would you have given to your younger self?”. This was the response:
I would tell my younger self,
- Force yourself to learn at least a bit of bioinformatics.
- Think big.
- You likely have at least some qualities, find them and make good use of them.
- Don’t wait for others to give you recognition, trust yourself, understand what you like, and have fun with it :)
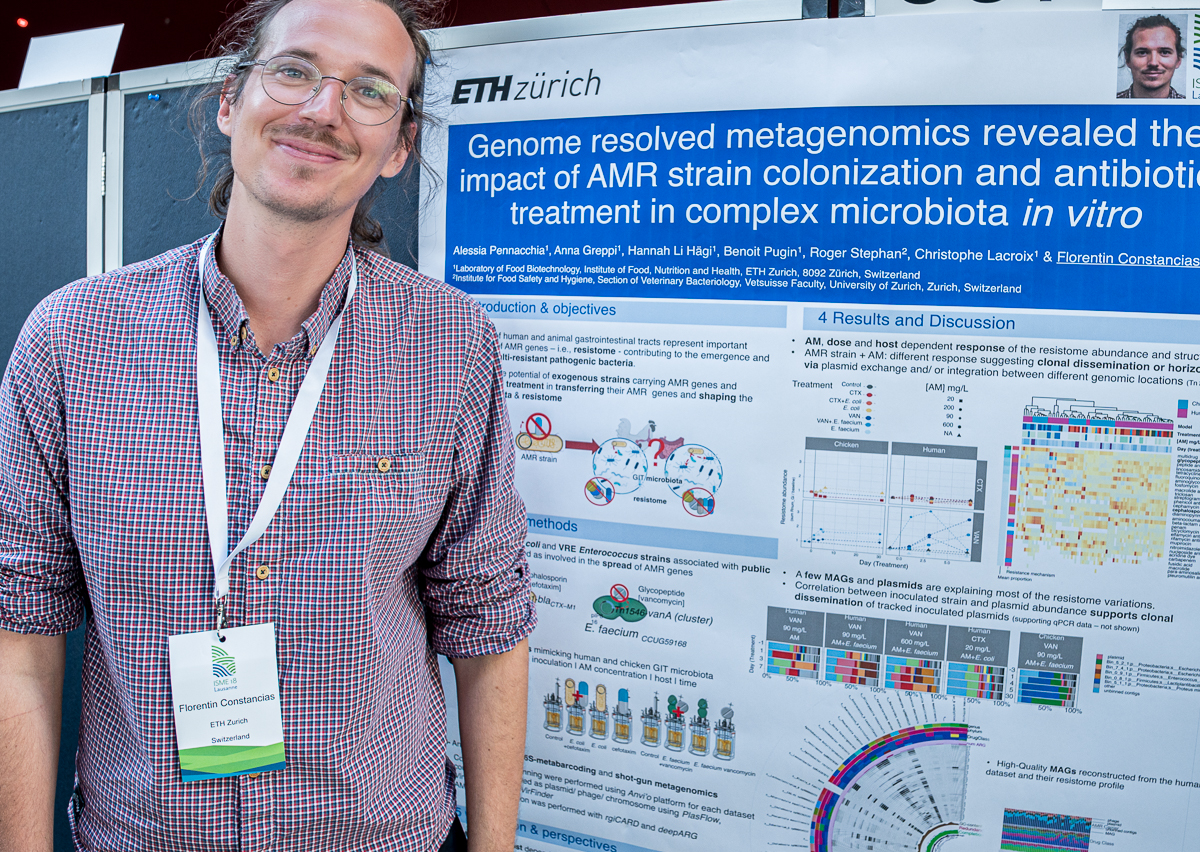
Florentin Constancias
Florentin’s poster at #ISME18 titled ‘Genome resolved metagenomics reveals the impact of AMR strain inoculation and antibiotic treatment in vitro’. When I asked what was the most interesting finding in this poster, Florentin said ‘We detected an host and antibiotic dependant response of the resistome and identified the population contributing to changes in resistome structure. We repeated the experiment to further confirm the robustness of the patterns and additional analysis are ongoing to identify potantial HGT events.’
Florentin Constancias (@fconstancias) is a computational microbial ecologist, and currently a postdoctoral scientist at the Laboratory of Food biotechnology at ETH Zurich, Switzerland. Florentin is generally interested in understanding the impact of antimicrobials on the human and animal gut resistome, the implication of microbiota in oral health and disease, and the recovery of metagenome-assembled genomes from ancient DNA to trace the genomic evolutionary history of oral bacteria.
Florentin has been an anvi’o user since version 2 :) We released v2 in 2016, and here is the release notes for those of you who would like to see “what was new” in anvi’o back then. As one of the very first users of anvi’o, Florentin has been using it primarily to reconstruct genomes from metagenomes. Florentin also mentioned that more recently he has been using SqueezeMeta pipeline to reconstruct MAGs, which includes an option to export the results to anvi’o. This enables Florentin to simultaneously extract gene centric as well as MAG-level information. He also said “it is quite satisfying when comparing the quality of the MAGs generated by default using the semi-automatic approach versus the manually curated one as recommended by anvi’o gurus! Just give it a try and you will see the difference!”. I asked Florentin “what do you like most about anvi’o (if you can think of anything you like about it)?” and this was his response:
Anvi’o is an amazing tool, supported by a very active and sharp community who paved the way for robust genome-resolved metagenomics for many of us and now contributes to research excellence and education while promoting full reproducibility: inspiring!
But,
Working on complex communities often requires combining approaches that enable a more exhaustive description of the microbiome than just MAGs. It would have been great to be able to combine genome resolved metagenomics and gene catalog approach within anvi’o to explore the -usually large- genetic signal which is not resolved by binning.
An important point. There are multiple very successful tools and approaches to generate gene catalogues (here is a critical assessment of a few), and it would have been excellent if anvi’o could help incorporating gene catalogues into its databases for integrated analyses.
If it was absolutely necessary to choose a different career path outside of research, Florentin would have chosen to be a farmer-baker or an ISME symposium organiser to keep in touch with the ISME community :). I also asked Florentin “given all the time you have spent in science, what advice would you have given to your younger self?”. This was his response:
Get things done!
An advice I constantly give my older self, too.
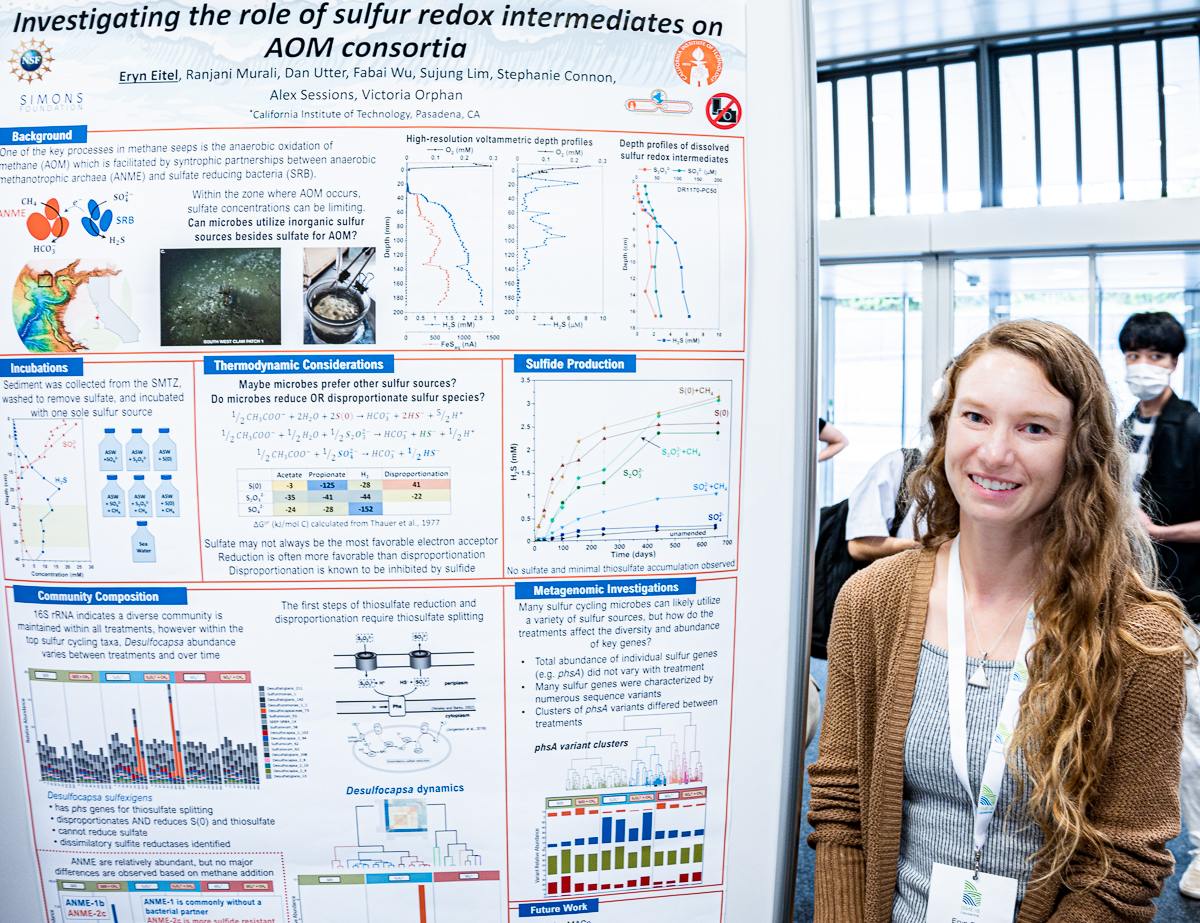
Eryn Eitel
Eryn’s poster at #ISME18 titled ‘Investigating the role of sulfur redox intermediates on AOM consortia’. When I asked what was the most interesting finding in this poster, Eryn said ‘although sulfate is generally more abundant in marine environments, microbial transformation of sulfur intermediate species, such as thiosulfate or elemental sulfur, may contribute to more rapid sulfide production in cold methane seeps.’
Eryn Eitel (@Aqueous_Eryn) is a biogeochemist (more specifically, a classical geochemist diving into microbial ecology), currently a postdoctoral scientist at the Sessions Lab as well as the Orphan Lab at the California Institute of Technology, United States, and generally interested in combining approaches to track transitory inorganic compounds with investigations of microbial community composition and activity to understand how environmental perturbations impact cryptic biogeochemical cycles.
Eryn has been an anvi’o user for less than 6 months, and has been using the platform primarily for processing metagenomic data from incubations to recover gene detection, function, and coverage patterns to compare between experiments. I asked Eryn “what do you like most about anvi’o (if you can think of anything you like about it)?”, and this was the response:
I like that it combines many aspects of the data together and puts everything in individual contigs-db files. This was a little difficult for me to grasp in the beginning, but I like it now that I understand that is what it is doing.
I also asked Eryn “what would you say about anvi’o to someone who never used it before to warn them?”
I know the anvi’o developers are trying to avoid turning anvi’o into a pipeline approach, but as an absolute beginner it was hard to understand what steps I could do in parallel versus what had to be sequential. I get it more now as I understand the nature of data products in anvi’o.
Also it would be nice to have either links to more information or more details about certain things for absolute beginners. For example it took me a little while to know why I should run both anvi-run-kegg-kofams and anvi-run-pfams, why are there two different anvi’o database files for the same project and how are they different? This makes more sense to me now that I have gone through more data, but I’m not sure if I could figure out much on my own from just the website tutorials without someone guiding me.
I know it is a hard balance to strike to reach both people that don’t know anything about metagenomics (or computers) and those that have used other methods before, but maybe just one or two extra pages or links to preexisting pages so that people who don’t know what is going on can have a little bit more to read or point towards other good resources, but everyone else can just skip those links. I also wish there was more contigs visualization options vs everything being mostly genome centered.
Great points, and I agree with all of them: anvi’o should do more to accommodate newcomers (especially from different disciplines), improve its documentation, and roll out new features that we have been working on for a long time to enable more in-depth comparative genomics beyond the visualization of individual contigs. I hope we will get there before alienating too many of the anvi’o users. With respect to documentation, we are thrilled to have contributions from those who walked some paths and would like to illuminate them for those who are only at the beginning of them. Perhaps we should start an F.A.Q. where people can submit all their questions anonymously, and then we can put together a community effort to organize those questions and write answers. It is indeed difficult to strike the right balance and be able to reach everyone, but researchers like Eryn (dealing with significant data challenges with their training in life sciences) are certainly within the center of what anvi’o aims to reach.
If it was absolutely necessary to choose a different career path outside of research, Eryn would have focused on furniture restoration :) I also asked Eryn “given all the time you have spent in science, what advice would you have given to your younger self?”:
Be better organized from the very beginning, and take better lab notes as you are doing things, not after the fact!
Igor Pessi
Igor’s poster at #ISME18 titled ‘Candidatus Nitrosopolaris, a genus of putative ammonia-oxidizing archaea with a polar/alpine distribution’. When I asked what was the most interesting finding in this poster, Igor said ‘looking at metagenomic data from northern Norway, we found a novel lineage of ammonia-oxidizing archaea that seems to have a very narrow geographic distribution limited to cold environments!’
Igor Pessi (@igor_spp) is a microbial ecologist, currently a postdoctoral scientist at Artic Microbial Ecology group of the University of Helsinki, Finland, and broadly interested in understanding how microbial communities that live in colder environments contribute to global ecosystem processes and how fragile they are to anthropogenic climate change.
Igor has been an anvi’o user for more than 2 years, and has been using the platform primarily for reconstructing genomes from metagenomes, phylogenomics, pangenomics, and eventually, in Igor’s own words, “plotting nice figures” :) I asked Igor’s take on the most positive aspects of anvi’o:
Hands down its philosophy of open-source, transparency, and the anvi’o user and developer community. All these make anvi’o such a dynamic tool that is constantly evolving.
I like the way anvi’o is structured, making it easy to integrate it with other tools. I also like the online tutorials and the funny error/warning messages :)
I also asked Igor what aspects of anvi’o could be considered challenging:
The learning curve can be somewhat steep, but the online tutorials and community resources make this path relatively easy.
If it was absolutely necessary to choose a different career path outside of research, Igor would have chosen to be a programmer. I also asked Igor “given all the time you have spent in science, what advice would you have given to your younger self?”:
Don’t take the academic career too seriously; learn the skills (technical or not) that you feel are important for you now and in the future, and keep an open mind about life outside of academia.
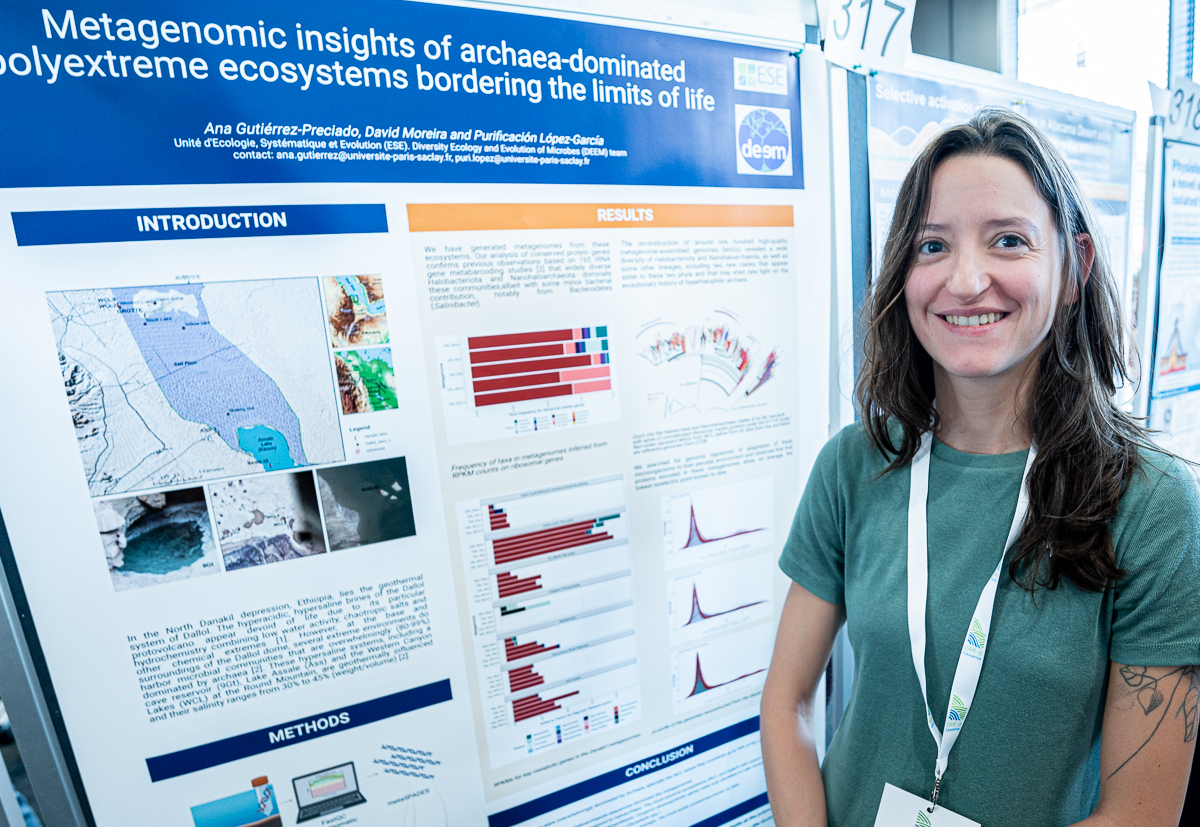
Ana Gutierrez-Preciado
Ana’s poster at #ISME18 titled ‘Metagenomic insights of archaea-dominated polyextreme ecosystems bordering the limits of life’. When I asked what was the most interesting finding in this poster, Ana said ‘(1) model organism are more the exception than the rule, and (2) space-for-time approaches can be applied to microbial communities to get insights on microbial metabolism on early Earth’
Ana Gutierrez-Preciado (@anagtz) is a bioinformatician and a microbiologist, currently a postdoctoral scientist with the DEEM Team at Universite Paris Saclay, and generally interested in understanding how microbes interact, evolve, thrive, and adapt to almost every environment.
Ana has been an anvi’o user for more than 2 years, and has been using the platform primarily through the metagenomics workflow with the aim of reconstructing genomes from metagenomes and for pangenomics. I asked Ana “what do you like most about anvi’o?” and got this response:
Every step is well studied, informed, and the best tools or options were carefully chosen for (at least) the metagenomics / metagenomic binning workflow.
I also like that when in doubt, I reached out to the community and they have always been quick and kind to respond and explain the details behind each step.
I like the versatility of anvi’o and the ability to incorporate different software or your own.
I also like all the online tutorials.
I also asked Ana’s take on what is missing in anvi’o:
I’m happy to leave this blank :) New users have a universe of tutorials on how to get started and a good community to back them up…
However… I also feel that a visual tool to explore the (comparative) genomic context of the newly reconstructed MAGs could be very useful for all of us users, and it could be very easily done integrating all the data from the anvi’o DBs. Something like this, but for anvi’o MAGs.
It’s been a long time we are aware of the fact that integrating all data into a genome visualization interface in anvi’o to interactively study biosynthetic gene clusters and or synteny of genes is a significant need. And almost everything is already in place, except for an interactive interface to give access to that. We have been working on it, and I am hopeful that there will be a solution sooner or later. We are also happy to implement ways to prepare data stored in anvi’o objects to quickly jump onto another tool to do this.
If it was absolutely necessary to choose a different career path outside of research, Ana would have chosen to be a nature photographer :) I also asked Ana “given all the time you have spent in science, what advice would you have given to your younger self?”. This was the response:
Always go for the challenge. It is worth it.
Antti Karkman
Antti’s poster at #ISME18 titled ‘Hosts and vehicles of antibiotic resistance genes in urban sewage’. Antti summarized the poster in a single sentence and said ‘antibiotic resistance genes in sewage are associated with a diverse set of mobile genetic elements and bacterial hosts.’
Antti Karkman (@anttikarkman) is a computational microbiologist, currently a Senior scientist at Molecular Environmental Biosciences Lab of the University of Helsinki, Finland, and generally interested in the environmental dimensions of antibiotic resistance.
Antti has been an anvi’o user for more than 2 years, and has been using the platform primarily for metagenomics and pangenomics. I asked Antti “what do you like most about anvi’o”:
The flexibility and number of different tools. And the network of scientists using and developing anvi’o.
Antti is a part of that network himself. The GFF3 parser Antti implemented for anvi’o has been very useful to many people who works with genomes annotated from Prokka or recovered from the IMG database.
I also want to share a separate memory from the conference that involves Antti: While visiting his poster, Antti showed me how the printing company failed to print the coverage of the contigs on it. While I quickly reached my black sharpie to rectify that error, Antti quickly reached his phone to immortalize that moment with a hilarious note:
Just tried the new anvi’o program, ’anvi-draw-contig-coverage’ and it works very well. #anvio #ISME18 pic.twitter.com/d6nTKDAbHN
— Antti Karkman (@AnttiKarkman) August 15, 2022
:)
Moritz Buck
Moritz’s poster at #ISME18 titled ‘mOTUpan: a robust Bayesian approach to leverage metagenome-assembled genomes for core-genome estimation’. When I asked what was the most interesting about this work, Moritz said ‘apparently if I am stuck in an embassy for 3 hours without a book or phone, I can finally understand Bayesian statistics’. I am jealous as I wish I had such positive experiences with embassies :/
Moritz Buck (@metamoritz) is a bioinformatician, currently a researcher at the Functional Microbial Ecology Group at the Swedish University of Agricultural Sciences, and generally interested in understanding how and why is microbial diversity shaped as it is.
Moritz has been a recent user of anvi’o and has been using the platform primarily for ad hoc interactive visualizations (i.e., using the program anvi-interactive with the --manual flag), but he also mentioned that he likes the general idea behind the platform so he tries to develop for anvi’o, at least to interface with it from his own programs. And indeed he did exactly that, and developed an anvi’o script, anvi-script-compute-bayesian-pan-core, that can run mOTUpan on a given anvi’o pangenome to identify the core set of genes in a Bayesian fashion, which is described here in this short and clear video.
During our exchange Moritz highlighted some of the serious shortcomings of anvi’o for programmers, such as its poorly documented API that poses a significant barrier for entry. While someone who is familiar with the codebase and data structures can do a lot of things without having to write a lot of code, anvi’o is as friendly to its new programmers as it is to its new users (minus the documentation the users could benefit from) :). As anvi’o developers we have been thinking a lot among ourselves about how to solve our needs of documentation for those who wish to use anvi’o from within their own programs with clear examples (like those shared at the end of this page). I hope we will see better days. But until proper documentation is ready, we are always happy to answer any developer questions to interface with anvi’o tools or data :)
If it was absolutely necessary to choose a different career path outside of research, Moritz would have chosen to be an engineer, photographer, brewer, chef, or literary-critic (or perhaps all of them at once!). I also asked Moritz “given all the time you have spent in science, what advice would you have given to your younger self?”:
Take more time before starting a PhD, learn how to work, and what you really like.
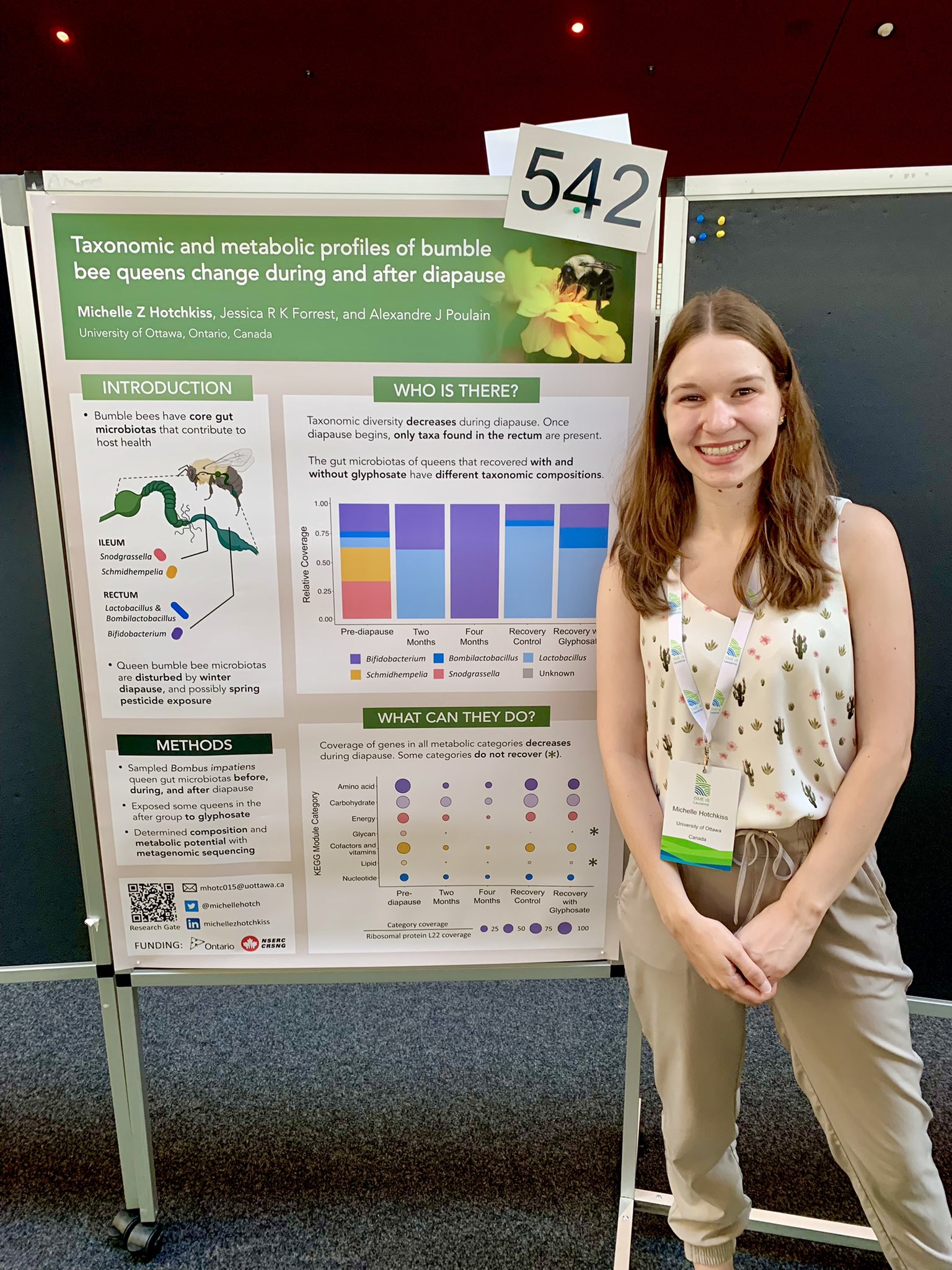
Michelle Z. Hotchkiss
Michelle’s poster at #ISME18 titled ‘Taxonomic and metabolic profiles of bumble bee queens change during and after diapause’. When I asked what was the most interesting finding in this poster, Michelle said ‘we found that all gram negative microbial taxa become undetectable after two months of diapause, and they remain undetected after one week of post-diapause recovery.’
Michelle Z. Hotchkiss (@michellehotch) is an entomologist and a microbiologist, currently a PhD candidate in the labs of Jessica Forrest and Alexandre Poulain at the University of Ottawa, Canada. Michelle is generally interested in understanding how various factors disturb the symbioses between native pollinators and their microbiota.
Michelle has been an anvi’o user for Less than 6 months, and has been using the platform primarily to process metagenomics data. I asked Michelle “what do you like most about anvi’o (if you can think of anything you like about it)?” and got this response:
The tutorials are easy to follow, and if I ever get stuck there’s a massive community of people to help.
I also asked Michelle “what would you say about anvi’o to someone who never used it before to warn them?”
Honestly nothing that I can think of.
If it was absolutely necessary to choose a different career path outside of research, Michelle would have chosen to be a lecturer. I also asked Michelle “given all the time you have spent in science, what advice would you have given to your younger self?”:
Schedule reoccurring mandatory fun for yourself (trivia night, running club, art class) that makes you take a break from science, have fun, and hang out with people outside of the lab. Your mental health will thank you!
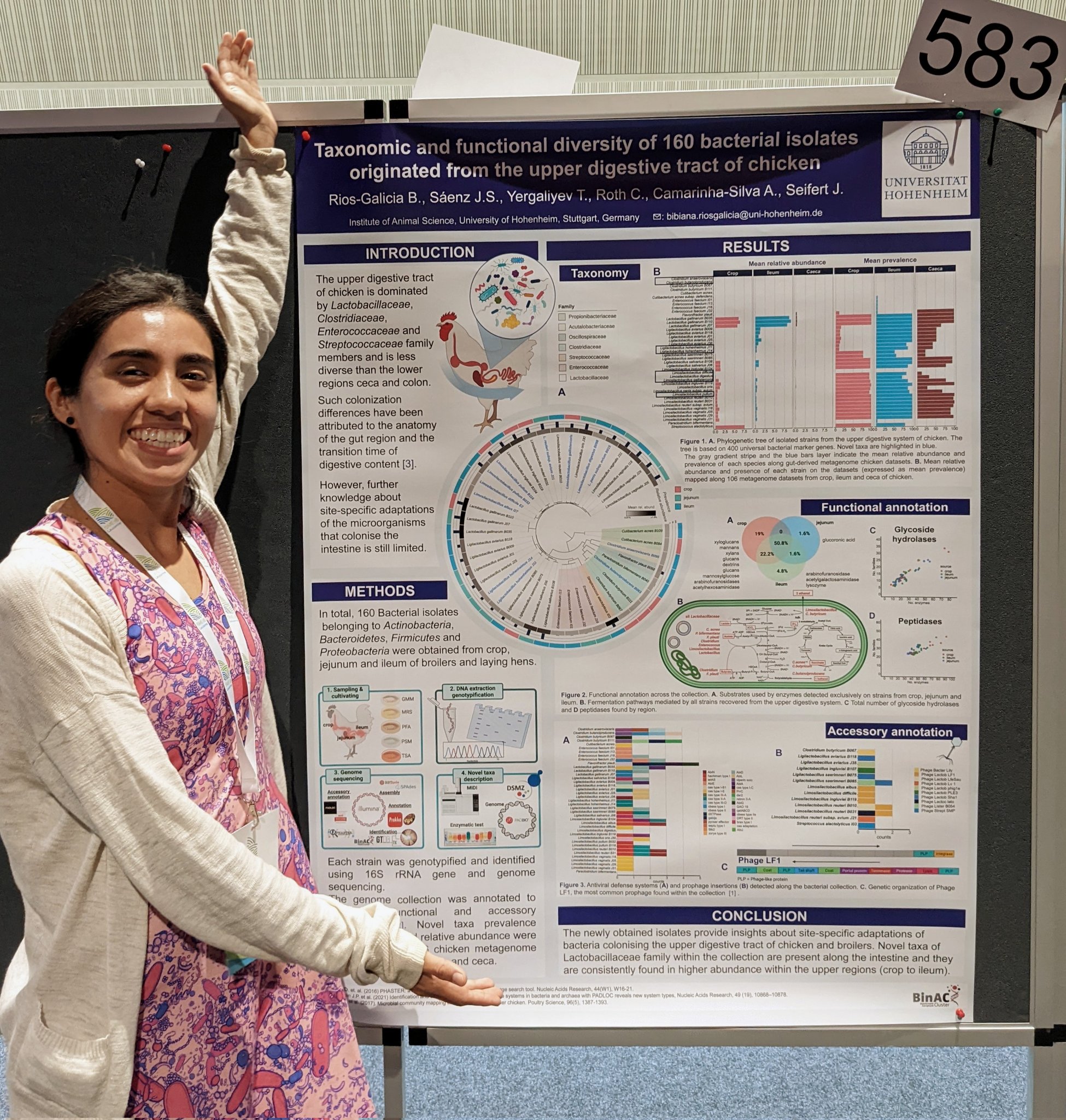
Bibiana Rios Galicia
Bibiana’s poster at #ISME18 titled ‘*Taxonomic and functional diversity of 160 bacterial isolates isolated originated from the upper digestive tract of chicken **’. When I asked what was the most interesting finding in this poster, Bibiana mentioned that they observed ‘different bacterial adaptations and community profiles throughout distinct regions of the digestive tract despite the short transition time of digestive content*’
Bibiana Rios Galicia (@Bibi_conBgorda) is a microbiologist and a microbe hunter :), currently a PhD candidate at Jana Seifert’s Group at the University of Hohenheim, Germany, and generally interested in understanding ways by which bacteria adapt to host anatomy.
Bibiana has been an anvi’o user for less than 6 months, and has been using the platform primarily for comparing genomes from the same species but different regions of isolation through pangenomics and phylogenomics. I asked Bibiana “what do you like most about anvi’o so far?” and got this response:
The feedback and comments in the terminal while running scripts, as well as the range of aesthetic possibilities in interactive interfaces.
I also asked Bibiana “what would you say about anvi’o to someone who never used it before to warn them?”
Remember to draw after every change to your plot, otherwise you don’t see your changes.
A problem we did everything in our power to address, and will continue to try to invent new ways to bring everyone’s attention to the mighty Draw button! :)
If it was absolutely necessary to choose a different career path outside of research, Bibiana would have chosen to be a baker. I also asked Bibiana “given all the time you have spent in science, what advice would you have given to your younger self?”:
When something simply does not work/run/make sense, and you don’t have a single clue what to change/improve, take a break, go eat something and come back. This time you will notice the mistake, and it works 100% of the time! :)
Which is exactly how I have been surviving my entire career! :)
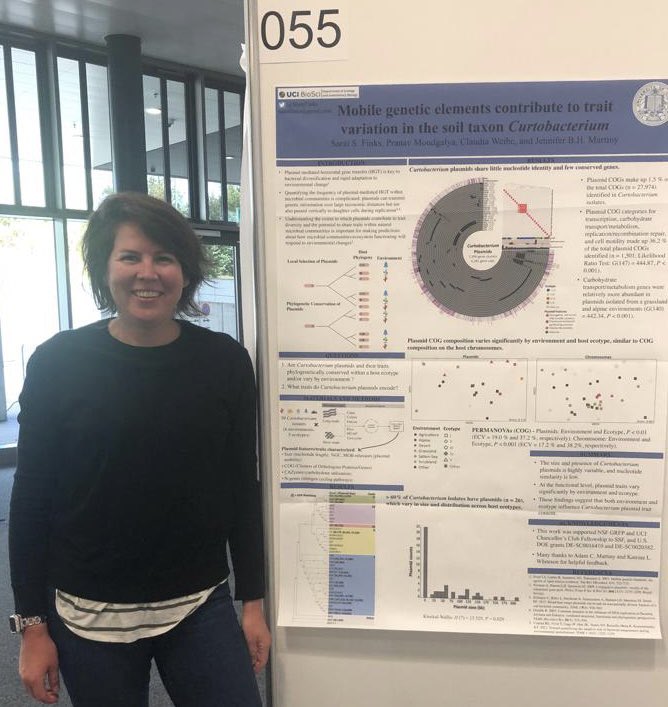
Sarai S. Finks
Sarai’s poster at #ISME18 titled ‘Mobile genetic elements contribute to trait variation in the soil taxon Curtobacterium’. When I asked what was the most interesting finding in this poster, Sarai said ‘about 1.5% of the 27,974 clusters of orthologous genes identified in Curtobacterium genomes were found on plasmids, which varied significantly in size and distribution across ecotypes .. more importantly, plasmid genes involved in carbon degradation (glycoside hydrolases) of plant litter and plasmid mobility were identified, suggesting these genes are potentially mobilizable within the bacterial communities where these Curtobacterium were isolated.’
Sarai S. Finks (@SaraiFinks) is a microbial ecologist and an evolutionary biologist, who is a newly minted Dr. from the Martiny Lab at the University of California, Irvine, soon to start in the Bordenstein Lab at the University of Penn State. Dr. Finks is interested in understanding how microbial communities adapt to environmental change, and how horizontal gene transfer events influence microbial communities functioning under different environmental conditions.
Sarai has been an anvi’o user for less than 2 years, but she has been doing a lot with it by feeding anvi’o with it by mostly performing pangenomic and phylogenomic analyses of hybrid genome assemblies (ONT long reads + Illumina short reads) of chromosomal and putative plasmid replicons together and separately, all in one place that includes (1) identifying gene clusters across replicon types, (2) estimating the relationships between/within replicon types based on gene clusters, (3) annotation of genes with COGs and PFAMs, (4) quantifying geometric homogeneity index of gene clusters to characterize within-gene-cluster alignment statistics, (5) calculating average nucleotide identity estimates for replicon types, (6) recovering concatenated single-copy core genes for phylogenomic analyses, (7) interactively visualizing replicon types to inspect amino acid alignments within gene clusters, (8) perform functional enrichment analyses on various groups (i.e., ecotypes verus environment), and (9) generating, in Sarai’s own words, “beautiful plots” :) Whew. By the power vested in me by the anvi’o developer community, I hereby pronounce Sarai, a “Champion of Anvi’o”, First of Her Name, Majesty of the Six Branches and Open PRs, and a Worthifier of the Realm. It is always an immensely gratifying experience for the developers to realize the diversity of ways researchers have put their efforts into use.
I asked Sarai “what do you like most about anvi’o?”:
I like that anvi’o has multiple workflows (some can be used in combination with one another) to wrangle and understand ‘omics data.
More importantly, I like that anvi’o is updated/maintained and there is a place to go to answer any questions that may arise on one’s anvi’o learning adventures (Slack and Tutorials :)).
I also like that if there is a useful tool for analyzing one’s data, the anvi’o team will add means to the codebase to export necessary files from anvi’o (e.g., to go into IQ-TREE).
Lastly, I like the Anvi’o interactive interface the most, it has been very helpful for me in understanding important aspects of my data and summarizing my findings in easy-to-interpret ways.
I also asked Sarai “what would you say about anvi’o to someone who never used it before to warn them?”:
Installing Anvi’o on a high performance cluster system and accessing the interactive display can be tricky, but with the right conda channel configuration and following along this tutorial, it can be done :)
Occasionally some of the terminology throws me, but of course all the vocabulary is nicely defined in https://anvio.org/vocabulary/.
I think Anvi’o was easy to learn, and I don’t feel it is impossible to keep up-to-date with what is going on, but maybe I don’t know what is going on :)
Also, if I have any questions, they are answered in the Slack channel and I can search through threads to see if anyone has encountered similar challenges and how they resolved them.
If it was absolutely necessary to choose a different career path outside of research, Sarai would have gone into science communication or become a science fiction writer (“in the same genre as Frank Herbert”, she noted). I also asked Sarai “given all the time you have spent in science, what advice would you have given to your younger self?”:
ASK MORE QUESTIONS and MAKE MORE MISTAKES. But not necessarily in this order.