anvi-run-kegg-kofams
Table of Contents
- Authors
- Can consume
- Can provide
- Usage
- How does it work?
- Standard usage
- Use a specific non-default KEGG data directory
- Run with multiple threads
- Use a different HMMER program
- Prevent HMMER from pre-filtering hits with default e-value reporting thresholds
- Keep all HMM hits
- Save the bitscores of HMM hits
- Modify the bitscore relaxation heuristic
- Skip BRITE annotations
- Annotate ‘No-threshold KOs’
- Additional Resources
Run KOfam HMMs on an anvi'o contigs database.
🔙 To the main page of anvi’o programs and artifacts.
Authors
Can consume
Can provide
Usage
Essentially, this program uses the KEGG database to annotate functions and metabolic pathways in a contigs-db. More specifically, anvi-run-kegg-kofams annotates a contigs-db with HMM hits from KOfam, a database of KEGG Orthologs (KOs). You must set up these HMMs on your computer using anvi-setup-kegg-data before you can use this program. If a modules-db is available, membership of KOfam functions in KEGG metabolic MODULES and BRITE hierarchies is also stored in the contigs-db.
Running this program is a pre-requisite for metabolism estimation with anvi-estimate-metabolism. Note that if you are planning to run metabolism estimation, it must be run with the same kegg-data that is used in this program to annotate KOfam hits.
How does it work?
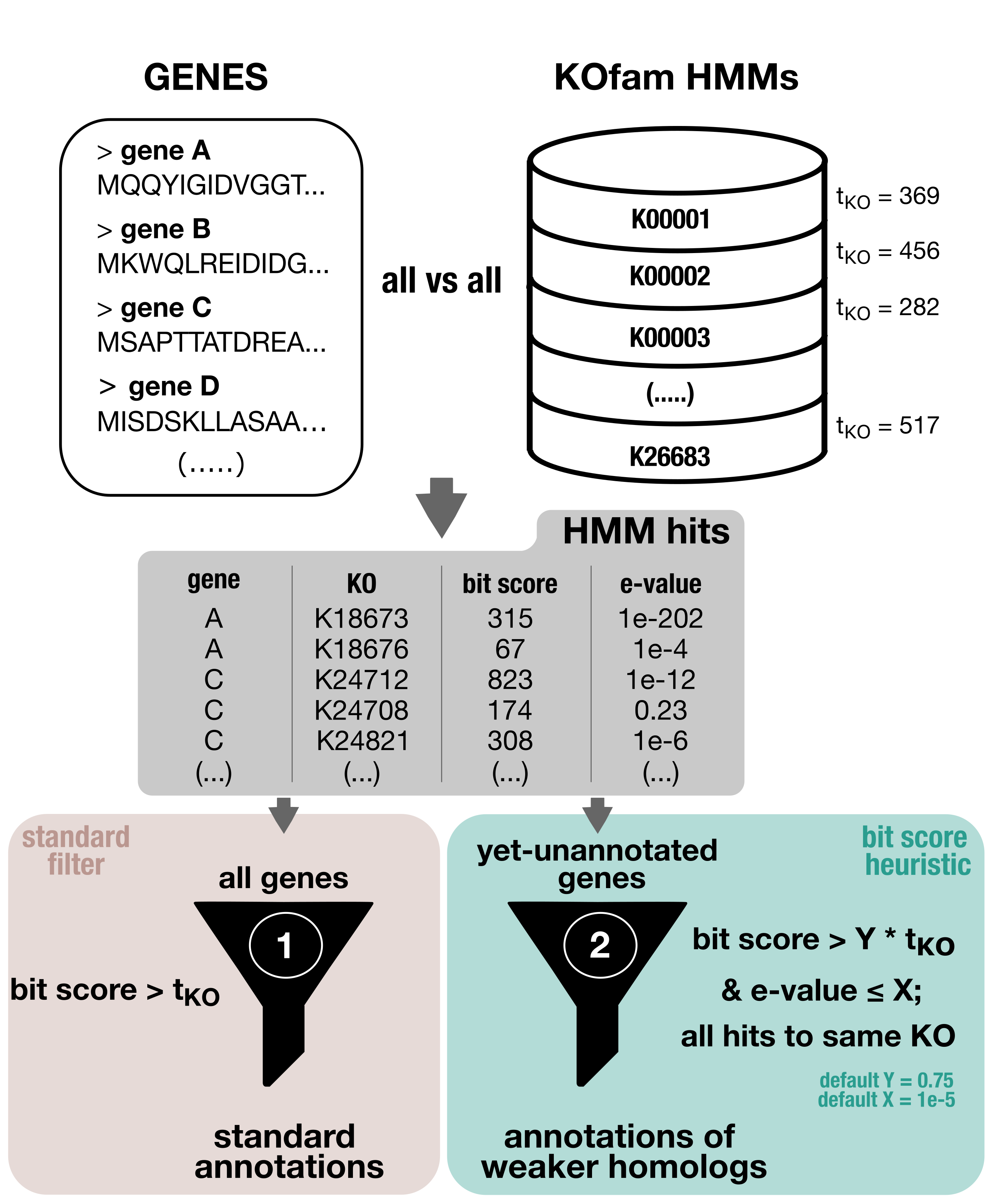
1) Run an HMM search against KOfam Briefly, what this program does is extract all the gene calls from the contigs-db and checks each one for hits to the KOfam HMM profiles in your kegg-data. This can be time-consuming given that the number of HMM profiles is quite large, even more so if the number of genes in the contigs-db is also large. Multi-threading is a good idea if you have the computational capability to do so.
2) Eliminate weak hits based on bitscore Many HMM hits will be found, most of them weak. The weak hits will by default be eliminated according to the bitscore thresholds provided by KEGG; that is, hits with bitscores below the threshold for a given KO profile will be discarded, and those with bitscores above the threshold will be annotated in the contigs-db. It is perfectly normal to notice that the number of raw hits found is many, many times larger than the number of annotated KO hits in your database.
3) Add back valid hits that were missed There is one issue with this practice of removing all KOfam hits below the KEGG bitscore threshold for a given profile. We (and others) have noticed that the KEGG thresholds can sometimes be too stringent, eliminating hits that are actually valid annotations. To solve this problem, we have implemented the following heuristic for relaxing the bitscore thresholds and annotating genes that would otherwise go without a valid KO annotation:
For every gene without a KOfam annotation, we examine all the hits with an e-value below X and a bitscore above Y percent of the threshold. If those hits are all to a unique KOfam profile, then we annotate the gene call with that KO.
X and Y are parameters that can be modified (see below), but by default the e-value threshold (X) is 1e-05 and the bitscore fraction (Y) is 0.75.
Please note that this strategy is just a heuristic. We have tried to pick default parameters that seemed reasonable but by no means have we comprehensively tested and optimized them. This is why X and Y are mutable so that you can explore different values and see how they work for your data. It is always a good idea to double-check your annotations to make sure they are reasonable and as stringent as you’d like them to be. In addition, if you do not feel comfortable using this heuristic at all, you can always turn this behavior off and rely solely on KEGG’s bitscore thresholds. :)
3) Put annotations in the database
In the contigs-db functions table, annotated KO hits (kegg-functions) will have the source KOfam. If a modules-db is available, metabolic modules and BRITE functional classifications containing these functions also have entries in the table, with sources labeled KEGG_Module and KEGG_BRITE. BRITE classification will not occur if anvi-setup-kegg-data was not set up with BRITE data (see the artifact for that program to see how to include BRITE).
Standard usage
anvi-run-kegg-kofams -c contigs-db
Use a specific non-default KEGG data directory
If you have previously setup your KEGG data directory using --kegg-data-dir (see anvi-setup-kegg-data), or have moved the KEGG data directory that you wish to use to a non-default location (maybe you like keeping the older versions around when you update, we don’t know how you roll), then you may need to specify where to find the KEGG data so that this program can use the right one. In that case, this is how you do it:
anvi-run-kegg-kofams -c contigs-db \ --kegg-data-dir /path/to/directory/KEGG
Run with multiple threads
anvi-run-kegg-kofams -c contigs-db -T 4
Use a different HMMER program
By default, anvi-run-kegg-kofams uses hmmsearch to find KO hits. If for some reason you would rather use a different program (hmmscan is also currently supported), you can do so.
anvi-run-kegg-kofams -c contigs-db \ --hmmer-program hmmscan
Prevent HMMER from pre-filtering hits with default e-value reporting thresholds
The default e-value reporting thresholds that HMMER uses are quite high, but e-values are dependent on the size of the databases used. So, if you have a very large dataset, it is possible that we miss some annotations from hits with bit scores that would be good enough to pass the bit score thresholds, but never get reported by HMMER in the first place because their e-values are too high. To avoid this, use the following flag:
anvi-run-kegg-kofams -c contigs-db \ --no-hmmer-prefiltering
In this case, we will set HMMER’s reporting thresholds to be extremely low and bit score-based (-T -20 and --domT -20) so that we don’t lose any hits with high bitscore yet high e-value. In most cases, this won’t lead to many new annotations because the bitscore filtering we apply later will weed out most raw hits. But please exercise caution. Using this flag with --keep-all-hits is not recommended.
Keep all HMM hits
Usually, this program parses out weak HMM hits and keeps only those that are above the score threshold for a given KO. If you would like to turn off this behavior and keep all hits (there will be a lot of weak ones), you can follow the example below:
anvi-run-kegg-kofams -c contigs-db \ --keep-all-hits
Save the bitscores of HMM hits
If you want to see the bitscores of all KOfam hits that were added to your contigs database, you can use the --log-bitscores option to save these values into a tab-delimited file:
anvi-run-kegg-kofams -c contigs-db \ --log-bitscores
Here is an example of what the resulting bitscore file would look like:
| entry_id | bit_score | domain_bit_score | e_value | entry_id | gene_callers_id | gene_hmm_id | gene_name |
|---|---|---|---|---|---|---|---|
| 1 | 177.4 | 85.1 | 8e-54 | 0 | 1371 | - | K10681 |
| 2 | 34.1 | 33.7 | 9.1e-11 | 1 | 1141 | - | K01954 |
| 3 | 22.4 | 22.4 | 3.1e-07 | 2 | 1402 | - | K01954 |
| 4 | 12.8 | 11.8 | 0.00024 | 3 | 1099 | - | K01954 |
| 5 | 17.1 | 16.7 | 4.4e-05 | 4 | 1267 | - | K20024 |
Combining this flag with the --keep-all-hits option is one way to get the bitscores of all matches to the KOfam profiles, even the ones that would usually not pass the bitscore threshold provided by KEGG.
Modify the bitscore relaxation heuristic
As described above, this program does its best to avoid missing valid annotations by relaxing the bitscore threshold for genes without any annotations. For such a gene, hits with e-value <= X and bitscore > (Y * KEGG threshold) that are all hits to the same KOfam profile are used to annotate the gene with that KO.
Skip this heuristic entirely
If you don’t want any previously-eliminated hits to be used for annotation, you can skip this heuristic by using the flag --skip-bitscore-heuristic. Then, only hits with bitscores above the KEGG-provided threshold for a given KO will be used for annotation.
anvi-run-kegg-kofams -c contigs-db \ --skip-bitscore-heuristic
Modify the heuristic parameters
If our default values are too stringent or not stringent enough for your tastes, you can change them! The e-value threshold (X, default: 1e-05) can be set using -E or --heuristic-e-value and the bitscore fraction (Y, default: 0.50) can be set using -H or --heuristic-bitscore-fraction. Like so:
anvi-run-kegg-kofams -c contigs-db \ -E 1e-15 \ -H 0.90
Skip BRITE annotations
If for some strange reason you do not want KEGG BRITE annotations to be added to your contigs database, you can skip them by providing the --skip-brite-hierarchies flag:
anvi-run-kegg-kofams -c contigs-db \ --skip-brite-hierarchies
Annotate ‘No-threshold KOs’
In every release of KOfam, there are some KO profiles that don’t come with predefined bit score thresholds. Normally, we skip annotating these. But if you want to annotate them, first read this documentation on our strategy for estimating bit score thresholds for nt-KOs so that you understand what is going on, and then run this program with the --include-nt-KOs flag:
anvi-run-kegg-kofams -c contigs-db \ --include-nt-KOs
Edit this file to update this information.
Additional Resources
Are you aware of resources that may help users better understand the utility of this program? Please feel free to edit this file on GitHub. If you are not sure how to do that, find the __resources__ tag in this file to see an example.

