anvi-estimate-metabolism [program]
Table of Contents
- Authors
- Can consume
- Can provide
- Usage
- Prerequisites to using this program
- Running metabolism estimation
- MULTI-MODE: Running metabolism estimation on multiple contigs databases
- Estimation for multiple single genomes
- Estimation for multiple bins
- Estimation for multiple metagenomes
- Adjustable Parameters
- Output options
- Testing this program
- Help! I’m getting version errors!
- Technical Details
- Additional Resources
Reconstructs metabolic pathways and estimates pathway completeness for a given set of contigs.
🔙 To the main page of anvi’o programs and artifacts.
Authors

Can consume
contigs-db ![]() kegg-data
kegg-data ![]() kegg-functions
kegg-functions ![]() profile-db
profile-db ![]() collection
collection ![]() bin
bin ![]() external-genomes
external-genomes ![]() internal-genomes
internal-genomes ![]() metagenomes
metagenomes ![]()
Can provide
Usage
anvi-estimate-metabolism predicts the metabolic capabilities of organisms based on their genetic content. It relies upon kegg-functions and metabolism information from KEGG (kegg-data), which is stored in a modules-db.
The metabolic pathways that this program currently considers are those defined by KEGG Orthologs (KOs) in the KEGG MODULE resource. Each KO represents a gene function, and a KEGG module is a set of KOs that collectively carry out the steps in a metabolic pathway.
Given a properly annotated contigs-db, this program determines which KOs are present and uses these functions to compute the completeness of each KEGG module. The output is one or more tabular text files - see kegg-metabolism for the output description and examples.
For a practical tutorial on how to use this program, visit this link. A more abstract discussion of available parameters, as well as technical details about how the metabolism estimation is done, can be found below.
Prerequisites to using this program
Metabolism estimation relies on gene annotations from the functional annotation source ‘KOfam’, also referred to as kegg-functions. Therefore, for this to work, you need to have annotated your contigs-db with hits to the KEGG KOfam database by running anvi-run-kegg-kofams prior to using this program.
Both anvi-run-kegg-kofams and anvi-estimate-metabolism rely on the kegg-data provided by anvi-setup-kegg-kofams, so if you do not already have that data on your computer, anvi-setup-kegg-kofams needs to be run first. To summarize, these are the steps that need to be done before you can use anvi-estimate-metabolism:
- Run anvi-setup-kegg-kofams to get data from KEGG onto your computer. This step only needs to be done once.
- Run anvi-run-kegg-kofams to annotate your contigs-db with kegg-functions. This program must be run on each contigs database that you want to estimate metabolism for.
Running metabolism estimation
You can run metabolism estimation on any set of annotated sequences, but these sequences typically fall under one of the following categories:
- Single genomes, also referred to as external-genomes. These can be isolate genomes or metagenome-assembled genomes, for example. Each one is described in its own individual contigs-db.
- Bins, also referred to as internal-genomes. These often represent metagenome-assembled genomes, but generally can be any subset of sequences within a database. A single contigs-db can contain multiple bins.
- Assembled, unbinned metagenomes. There is no distinction between sequences that belong to different microbial populations in the contigs-db for an unbinned metagenome.
As you can see, anvi-estimate-metabolism always takes one or more contigs database(s) as input, but the information that is taken from those databases depends on the context (ie, genome, metagenome, bin). In the case of internal genomes (or bins), is possible to have multiple inputs but only one input contigs db. So for clarity’s sake, we sometimes refer to the inputs as ‘samples’ in the descriptions below. If you are getting confused, just try to remember that a ‘sample’ can be a genome, a metagenome, or a bin.
Different input contexts can require different parameters or additional inputs. The following sections describe what is necessary for each input type.
Estimation for a single genome
The most basic use-case for this program is when you have one contigs database describing a single genome. Since all of the sequences in this database belong to the same genome, all of the gene annotations will be used for metabolism estimation.
anvi-estimate-metabolism -c contigs-db
Estimation for bins in a metagenome
You can estimate metabolism for different subsets of the sequences in your contigs database if you first bin them and save them as a collection. For each bin, only the gene annotations from its subset of sequences will contribute to the module completeness scores.
You can estimate metabolism for every individual bin in a collection by providing the profile database that describes the collection as well as the collection name:
anvi-estimate-metabolism -c contigs-db -p profile-db -C collection
The metabolism estimation results for each bin will be printed to the same output file(s). The bin_name column in long-format output will distinguish between results from different bins.
If you only want estimation results for a single bin, you can instead provide a specific bin name from that collection using the -b parameter:
anvi-estimate-metabolism -c contigs-db -p profile-db -C collection -b bin
Or, to estimate on a subset of bins in the collection, you can provide a text file containing the specific list of bins that you are interested in:
anvi-estimate-metabolism -c contigs-db -p profile-db -C collection -B bin_ids.txt
Each line in the bin_ids.txt file should be a bin name from the collection (there is no header line). Here is an example file containing three bin names:
bin_1
bin_3
bin_5
Estimation for a metagenome
If you have an unbinned metagenome assembly, you can estimate metabolism for it using --metagenome-mode. In this case, since there is no way to determine which contigs belong to which microbial populations in the sample, estimation will be done on a per-contig basis; that is, for each contig, only the genes present on that contig will be used to determine pathway completeness within the contig.
anvi-estimate-metabolism -c contigs-db --metagenome-mode
In metagenome mode, this program will estimate metabolism for each contig in the metagenome separately. This will tend to underestimate module completeness because it is likely that many modules will be broken up across multiple contigs belonging to the same population. If you prefer to instead treat all KOs in the metagenome as belonging to one collective genome, you can do so by simply leaving out the --metagenome-mode flag (to effectively pretend that you are doing estimation for a single genome, although in your heart you will know that your contigs database really contains a metagenome). Please note that this will result in the opposite tendency to overestimate module completeness (as the KOs will in reality be coming from multiple different populations), and there will be a lot of redundancy. We are working on improving our estimation algorithm for metagenome mode. In the meantime, if you are worried about the misleading results from either of these situations, we suggest binning your metagenomes first and running estimation for the bins as described below.
MULTI-MODE: Running metabolism estimation on multiple contigs databases
If you have a set of contigs databases of the same type (i.e., all of them are single genomes or all are binned metagenomes), you can analyze them all at once. What you need to do is put the relevant information for each contigs-db into a text file and pass that text file to anvi-estimate-metabolism. The program will then run estimation individually on each contigs database in the file. The estimation results for each database will be aggregated and printed to the same output file(s).
One advantage that multi-mode unlocks is the ability to generate matrix-formatted output, which is convenient for clustering or visualizing the metabolic potential of multiple samples. See the Output options section below for more details.
Estimation for multiple single genomes
Multiple single genomes (also known as external-genomes) can be analyzed with the same command by providing an external genomes file to anvi-estimate-metabolism. To see the required format for the external genomes file, see external-genomes.
anvi-estimate-metabolism -e external-genomes.txt
Estimation for multiple bins
If you have multiple bins (also known as internal-genomes), they can be analyzed with the same command by providing an internal genomes file to anvi-estimate-metabolism. The bins in this file can be from the same collection, from different collections, or even from different metagenomes. To see the required format for the internal genomes file, see internal-genomes.
anvi-estimate-metabolism -i internal-genomes.txt
Estimation for multiple metagenomes
Multiple metagenomes can be analyzed with the same command by providing a metagenomes input file. Metagenome mode will be used to analyze each contigs database in the file. To see the required format for the metagenomes file, see metagenomes.
anvi-estimate-metabolism -M metagenomes.txt
Adjustable Parameters
There are many ways to alter the behavior of this program to fit your needs. You can find some commonly adjusted parameters below. For a full list of parameters, check the program help (-h) output.
Changing the module completion threshold
As explained in the technical details section below, KEGG module completeness is computed as the percentage of steps in the metabolic pathway that are ‘present’ based on the annotated KOs in the contigs database. If this completeness is larger than a certain percentage, then the entire module is considered to be ‘complete’ in the sample and the corresponding row in the long-format modules mode output file will have ‘True’ under the module_is_complete column. By default, the module completion threshold is 0.75, or 75%.
Changing this parameter doesn’t have any effect other than changing the proportions of ‘True’ and ‘False’ values in the module_is_complete column of long-format modules mode output (or the proportion of 1s and 0s in the module presence-absence matrix for --matrix-format output). It does not alter completeness scores. It also does not affect which modules are printed to the output file, unless you use the --only-complete flag (described in a later section). Therefore, the purpose of changing this threshold is usually so that you can filter the output later somehow (i.e., by searching for ‘True’ values in the long-format output).
In this example, we change the threshold to 50 percent.
anvi-estimate-metabolism -c contigs-db --module-completion-threshold 0.5
Working with a non-default KEGG data directory
If you have previously annotated your contigs databases using a non-default KEGG data directory with --kegg-data-dir (see anvi-run-kegg-kofams), or you have moved the KEGG data directory that you wish to use to a non-default location, then you will need to specify where to find the KEGG data so that this program can use the right one. In that case, this is how you do it:
anvi-estimate-metabolism -c contigs-db --kegg-data-dir /path/to/directory/KEGG
Output options
This program has two types of output files: long-format (tab-delimited) output files and matrices. The long-format output is the default. If you are using multi-mode to work with multiple samples, you can request matrix output by using the flag --matrix-format.
You can find more details on the output formats by looking at kegg-metabolism. Below, you will find examples of how to use output-related parameters.
Long-Format Output
Long-format output has several preset “modes” as well as a “custom” mode in which the user can define the contents of the output file. Multiple modes can be used at once, and each requested “mode” will result in a separate output file. The default output mode is “modules” mode.
Viewing available output modes
To see what long-format output modes are currently available, use the --list-available-modes flag:
anvi-estimate-metabolism -c contigs-db --list-available-modes
The program will print a list like the one below and then exit.
AVAILABLE OUTPUT MODES
===============================================
kofam_hits_in_modules ........................: Information on each KOfam hit that belongs to a KEGG module
modules ......................................: Information on KEGG modules
modules_custom ...............................: A custom tab-delimited output file where you choose the included KEGG modules data using --custom-output-headers
kofam_hits ...................................: Information on all KOfam hits in the contigs DB, regardless of KEGG module membership
Please note that you must provide your input file(s) for this to work.
Using a non-default output mode
You can specify one or more long-format output modes using the --kegg-output-modes parameter. The mode names must exactly match to one of the available modes from the --list-available-modes output.
anvi-estimate-metabolism -c contigs-db --kegg-output-modes kofam_hits
Using multiple output modes
If you want more than one output mode, you can provide multiple comma-separated mode names to the --kegg-output-modes parameter. There should be no spaces between the mode names.
anvi-estimate-metabolism -c contigs-db --kegg-output-modes kofam_hits,modules
When multiple output modes are requested, a different output file is produced for each mode. All output files will have the same prefix, and the file suffixes specify the output mode. For example, modules mode output has the suffix _modules.txt while kofam hits mode has the suffix _kofam_hits.txt.
Viewing available output headers for ‘custom’ mode
The modules_custom output mode allows you to specify which information to include (as columns) in your long-format output. It is essentially a customizable version of modules mode output. To use this mode, you must specify which columns to include by listing the column names after the --custom-output-headers flag.
To find out what column headers are available, use the --list-available-output-headers parameter:
anvi-estimate-metabolism -c contigs-db --list-available-output-headers
The program will print a list like the one below and then exit.
AVAILABLE OUTPUT HEADERS
===============================================
unique_id ....................................: Just an integer that keeps our data organized. No real meaning here. Always included in output, so no need to specify it on the command line [all output modes]
kegg_module ..................................: KEGG module number [modules output mode]
module_is_complete ...........................: Whether a KEGG module is considered complete or not based on its percent completeness and the completeness threshold [modules output mode]
module_completeness ..........................: Percent completeness of a KEGG module [modules output mode]
module_name ..................................: English name/description of a KEGG module [modules output mode]
module_class .................................: Metabolism class of a KEGG module [modules output mode]
module_category ..............................: Metabolism category of a KEGG module [modules output mode]
module_subcategory ...........................: Metabolism subcategory of a KEGG module [modules output mode]
module_definition ............................: KEGG-formatted definition of a KEGG module. Describes the metabolic pathway in terms of the KOS that belong to the module [modules output mode]
module_substrates ............................: Comma-separated list of compounds that serve as initial input to the metabolic pathway (that is, substrate(s) to the initial reaction(s) in the pathway) [modules output mode]
module_products ..............................: Comma-separated list of compounds that serve as final output from the metabolic pathway (that is, product(s) of the final reaction(s) in the pathway) [modules output mode]
module_intermediates .........................: Comma-separated list of compounds that are intermediates the metabolic pathway (compounds that are both outputs and inputs of reaction(s) in the pathway) [modules output mode]
gene_caller_ids_in_module ....................: Comma-separated list of gene caller IDs of KOfam hits in a module [modules output mode]
gene_caller_id ...............................: Gene caller ID of a single KOfam hit in the contigs DB. If you choose this header, each line in the output file will be a KOfam hit [all output modes]
kofam_hits_in_module .........................: Comma-separated list of KOfam hits in a module [modules output mode]
kofam_hit ....................................: KO number of a single KOfam hit. If you choose this header, each line in the output file will be a KOfam hit [modules output mode]
contig .......................................: Contig that a KOfam hit is found on. If you choose this header, each line in the output file will be a KOfam hit [all output modes]
path_id ......................................: Integer ID for a path through a KEGG module. No real meaning and just for data organization. If you choose this header, each line in the output file will be a KOfam hit
[modules output mode]
path .........................................: A path through a KEGG module (a linear sequence of KOs that together represent each metabolic step in the module. Most modules have several of these due to KO redundancy). If
you choose this header, each line in the output file will be a KOfam hit [modules output mode]
path_completeness ............................: Percent completeness of a particular path through a KEGG module. If you choose this header, each line in the output file will be a KOfam hit [modules output mode]
warnings .....................................: If we are missing a KOfam profile for one of the KOs in a module, there will be a note in this column. [modules output mode]
ko ...........................................: KEGG Orthology (KO) number of a KOfam hit [kofams output mode]
modules_with_ko ..............................: A comma-separated list of modules that the KO belongs to [kofams output mode]
ko_definition ................................: The functional annotation associated with the KO number [kofams output mode]
genome_name ..................................: Name of genome/bin/metagenome in which we find KOfam hits and/or KEGG modules [all output modes]
As you can see, this flag is also useful when you want to quickly look up the description of each column of data in your output files.
For each header, the output mode(s) that it is applicable to are listed after the description. The headers you can choose from for modules_custom output end in either [modules output mode] or [all output modes].
Just as with --list-available-modes, you must provide your input file(s) for this to work. In fact, some headers will change depending on which input types you provide.
Using custom output mode
Here is an example of defining the modules output to contain columns with the module number, the module name, and the completeness score. The corresponding headers for these columns are provided as a comma-separated list (no spaces) to the --custom-output-headers flag.
anvi-estimate-metabolism -c contigs-db --kegg-output-modes modules_custom --custom-output-headers kegg_module,module_name,module_completeness
Including modules with 0% completeness in long-format output
By default, modules with a completeness score of 0 are not printed to the output files to save on space. But you can explicitly include them by adding the --include-zeros flag.
anvi-estimate-metabolism -c contigs-db --include-zeros
Including coverage and detection in long-format output
If you have a profile database associated with your contigs database and you would like to include coverage and detection data in the metabolism estimation output files, you can use the --add-coverage flag. You will need to provide the profile database as well, of course. :)
anvi-estimate-metabolism -c contigs-db -p profile-db --kegg-output-modes modules,kofam_hits_in_modules,kofam_hits --add-coverage
For kofam_hits_in_modules and kofam_hits mode output files, in which each row describes one KOfam hit to a gene in the contigs database, the output will contain two additional columns per sample in the profile database. One column will contain the mean coverage of that particular gene call by reads from that sample and the other will contain the detection of that gene in the sample.
For modules mode output files, in which each row describes a KEGG module, the output will contain four additional columns per sample in the profile database. One column will contain comma-separated mean coverage values for each gene call in the module, in the same order as the corresponding gene calls in the gene_caller_ids_in_module column. Another column will contain the average of these gene coverage values, which represents the average coverage of the entire module. Likewise, the third and fourth columns will contain comma-separated detection values for each gene call and the average detection, respectively.
Note that you can customize which coverage/detection columns are in the output files if you use custom modules mode. Use the following command to find out which coverage/detection headers are available:
anvi-estimate-metabolism -c contigs-db -p profile-db --add-coverage --list-available-output-headers
Matrix Output
Matrix format is only available when working with multiple contigs databases. Several output matrices will be generated, each of which describes one statistic such as module completion score, module presence/absence, or KO hit counts. As with long-format output, each output file will have the same prefix and the file suffixes will indicate which type of data is present in the file.
In each matrix, the rows will describe modules or KOs, the columns will describe your input samples (i.e. genomes, metagenomes, bins), and each cell will be the corresponding statistic. You can see examples of this output format by viewing kegg-metabolism.
Obtaining matrix-formatted output
Getting these matrices is as easy as providing the --matrix-format flag.
anvi-estimate-metabolism -i internal-genomes.txt --matrix-format
Including KEGG metadata in the matrix output
By default, the matrix output is a matrix ready for use in other computational applications, like visualizing as a heatmap or performing clustering. That means it has a header line and an index in the right-most column, but all other cells are numerical. However, you may want to instead have a matrix that is annotated with more information, like the names and categories of each module or the functional annotations of each KO. To include this additional information in the matrix output (as columns that occur before the sample columns), use the --include-metadata flag.
anvi-estimate-metabolism -i internal-genomes.txt --matrix-format --include-metadata
Note that this flag only works for matrix output because, well, the long-format output inherently includes KEGG metadata.
Including rows of all zeros in the matrix output
The --include-zeros flag works for matrix output, too. By default, modules that have 0 completeness (or KOs that have 0 hits) in every input sample will be left out of the matrix files. Using --include-zeros results in the inclusion of these items (that is, the inclusion of rows of all zeros).
anvi-estimate-metabolism -i internal-genomes.txt --matrix-format --include-zeros
Getting module-specific KO hit matrices
The standard KO hit matrix includes all KOfams that were annotated at least once in your input databases (or all KOfams that we know about, if --include-zeros is used). But sometimes you might want to see a matrix with only the KOs from a particular metabolic pathway. To do this, pass a comma-separated (no spaces) list of KEGG module numbers to the --module-specific-matrices flag, and then your matrix output will include KO hit matrices for each module in the list.
For example,
anvi-estimate-metabolism -e input_txt_files/external_genomes.txt \ --matrix-format \ --module-specific-matrices M00001,M00009 \ -O external_genomes
will produce the output files external_genomes-M00001_ko_hits-MATRIX.txt and external_genomes-M00009_ko_hits-MATRIX.txt (in addition to the typical output matrices). Each additional output matrix will include one row for each KO in the module, in the order it appears in the module definition. It will also include comment lines for each major step (or set of steps) in the module definition, to help with interpreting the output.
Check out this (partial) example for module M00001:
KO isolate E_faecalis_6240 test_2
# (K00844,K12407,K00845,K00886,K08074,K00918)
K00844 0 0 0
K12407 0 0 0
K00845 0 1 0
K00886 1 0 1
K08074 0 0 0
K00918 0 0 0
# (K01810,K06859,K13810,K15916)
K01810 1 1 1
K06859 0 0 0
K13810 0 0 0
K15916 0 0 0
# (K00850,K16370,K21071,K00918)
K00850 0 1 0
K16370 0 0 0
K21071 0 0 0
K00918 0 0 0
[....]
If you don’t want those comment lines in there, you can combine this with the --no-comments flag to get a clean matrix. This might be useful if you want to do some downstream processing of the matrices.
anvi-estimate-metabolism -e input_txt_files/external_genomes.txt \ --matrix-format \ --module-specific-matrices M00001,M00009 \ --no-comments \ -O external_genomes
In this case, the above file would look like this:
KO isolate E_faecalis_6240 test_2
K00844 0 0 0
K12407 0 0 0
K00845 0 1 0
K00886 1 0 1
K08074 0 0 0
K00918 0 0 0
K01810 1 1 1
K06859 0 0 0
K13810 0 0 0
K15916 0 0 0
K00850 0 1 0
K16370 0 0 0
K21071 0 0 0
K00918 0 0 0
[....]
Other output options
Regardless of which output type you are working with, there are a few generic options for controlling how the output files look like.
Changing the output file prefix
anvi-estimate-metabolism can produce a variety of output files. All will be prefixed with the same string, which by default is kegg-metabolism. If you want to change this prefix, use the -O flag.
anvi-estimate-metabolism -c contigs-db -O my-cool-prefix
Including only complete modules in the output
Remember that module completion threshold? Well, you can use that to control which modules make it into your output files. If you provide the --only-complete flag, then any module-related output files will only include modules that have a completeness score at or above the module completion threshold. (This doesn’t affect KO-related outputs, for obvious reasons.)
Here is an example of using this flag with long-format output (which is the default, as described above, but we are asking for it explicitly here just to be clear):
anvi-estimate-metabolism -c contigs-db --kegg-output-modes modules --only-complete
And here is an example of using this flag with matrix output. In this case, we are working with multiple input samples, and the behavior of this flag is slightly different: a module will be included in the matrix if it is at or above the module completion threshold in at least one sample. If there are any samples in which that module’s completeness is below the threshold, its completeness in that sample will be represented by a 0.0 in the matrix, regardless of its actual completeness score.
anvi-estimate-metabolism -i internal-genomes.txt --matrix-format --only-complete
Testing this program
You can see if this program is working on your computer by running the following suite of tests, which will check several common use-cases:
anvi-self-test --suite metabolism
Help! I’m getting version errors!
If you have gotten an error that looks something like this:
Config Error: The contigs DB that you are working with has been annotated with a different version of the MODULES.db than you are working with now.
This means that the modules-db used by anvi-run-kegg-kofams has different contents (different KOs and/or different modules) than the one you are currently using to estimate metabolism, which would lead to mismatches if metabolism estimation were to continue. There are a few ways this can happen, which of course have different solutions:
- You annotated your contigs-db with a former version of kegg-data, and subsequently set up a new anvi-setup-kegg-kofams (possibly with the
--kegg-archiveor--download-from-keggoptions, which get you a non-default version of KEGG data). Then you tried to run anvi-estimate-metabolism with the new kegg-data version. If this is you, and you have saved your former version of kegg-data somewhere, then you are in luck - you can simply direct anvi-estimate-metabolism to use the old version of KEGG with--kegg-data-dir. If you didn’t save it, then unfortunately you will most likely have to re-run anvi-run-kegg-kofams on your contigs-db to re-annotate it with the new version before continuing with metabolism estimation. - You have multiple versions of kegg-data on your computer in different locations, and you used different ones for anvi-run-kegg-kofams and anvi-estimate-metabolism. If this is what you did, then there is an easy fix - simply find the KEGG data directory containing the MODULES.db with the same content hash (you can use anvi-db-info on the MODULES.db to find the hash value) as the one used by anvi-run-kegg-kofams and submit that location with
--kegg-data-dirto this program. - Your collaborator gave you some databases that they annotated with a different version of kegg-data than you have on your computer. In this case, either you or they (or both) have probably been using a non-default (or outdated) version of kegg-data. If they have the current default snapshot of KEGG data but you do not, then you’ll need to get that version onto your computer using the default usage of anvi-setup-kegg-kofams. Otherwise, your collaborator will need to somehow share all or part of their KEGG data directory with you before you can work on their databases. See anvi-setup-kegg-kofams for details on how to share non-default setups of kegg-data.
Technical Details
What data is used for estimation?
Regardless of which input type is provided to this program, the basic requirements for metabolism estimation are 1) a set of metabolic pathway definitions, and 2) a ‘pool’ of gene annotations.
Module Definitions
Currently, the only option for metabolic pathway definitions that is available to this program is the KEGG MODULE resource. The program anvi-setup-kegg-kofams acquires the definitions of these modules using the KEGG API and puts them into the modules-db. The definitions are strings of KEGG Ortholog (KO) identifiers, representing the functions necessary to carry out each step of the metabolic pathway. Let’s use module M00018, Threonine Biosynthesis, as an example. Here is the module definition, in picture form:
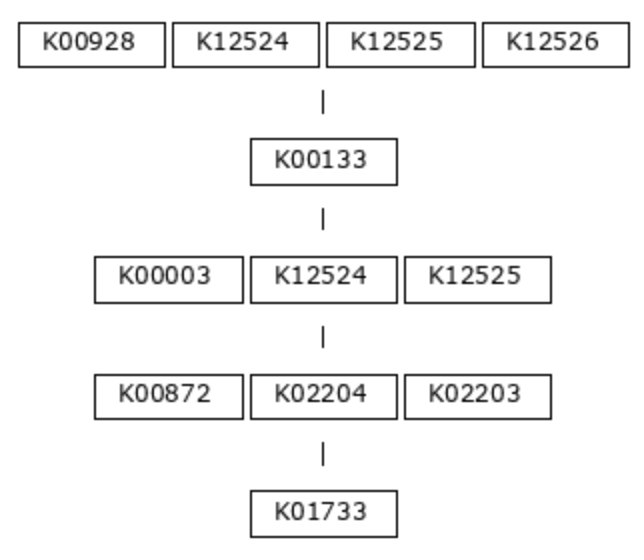
This biosynthesis pathway has five steps, or chemical reactions. The first reaction in the pathway requires an aspartate kinase enzyme (also known as a homoserine dehydrogenase), and there are four possible orthologs known to encode this function: K00928, K12524, K12525, or K12526. Only one of these genes is required to be able to carry out this step. In contrast, the second reaction can be fulfilled by only one known KO, the aspartate-semialdehyde dehydrogenase K00133.
The definition string for module M00018 is this:
(K00928,K12524,K12525,K12526) K00133 (K00003,K12524,K12525) (K00872,K02204,K02203) K01733
Hopefully the correspondence between the picture and text is clear - spaces separate distinct steps in the pathway, while commas separate alternatives.
That was a simple example, so let’s look at a more complicated one: M00011, the second carbon oxidation phase of the citrate cycle.
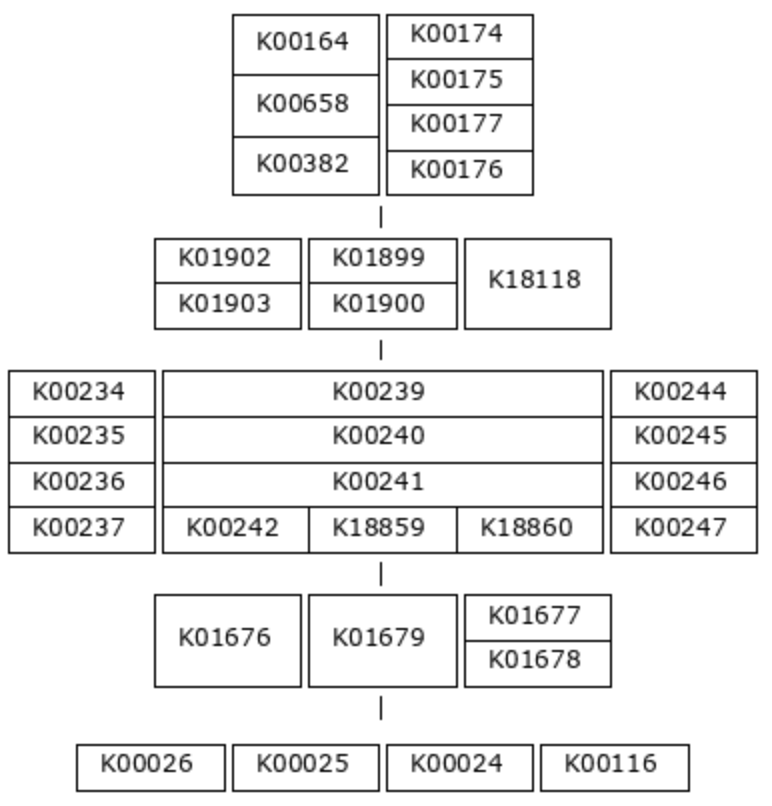
This pathway also has five steps, but this time, most of the reactions require an enzyme complex. Each KO within a multi-KO box is a component of an enzyme. For example, one option for the first reaction is 2-oxoglutarate dehydrogenase, a 3-component enzyme made up of K00164, K00658, and K00382.
This is the definition string for module M00011:
(K00164+K00658+K00382,K00174+K00175-K00177-K00176) (K01902+K01903,K01899+K01900,K18118) (K00234+K00235+K00236+K00237,K00239+K00240+K00241-(K00242,K18859,K18860),K00244+K00245+K00246-K00247) (K01676,K01679,K01677+K01678) (K00026,K00025,K00024,K00116)
And here is a detail that is difficult to tell from the pictorial definition - not all enzyme components are equally important. You can see in the definition string that KO components of an enzyme are connected with either ‘+’ signs or ‘-‘ signs. The ‘+’ sign indicates that the following KO is an essential component of the enzyme, while the ‘-‘ sign indicates that it is non-essential. For the purposes of module completeness estimation, we only consider a reaction to be fulfilled if all the essential component KOs are present in the annotation pool (and we don’t care about the ‘non-essential’ components). So, for example, we would consider the first step in this pathway complete if just K00174 and K00175 were present. The presence/absence of either K00177 or K00176 would not affect the module completeness score at all.
Module definitions can be even more complex than this. Both of these examples had exactly five steps, no matter which set of KOs you use to fulfill each reaction. However, in some modules, there can be alternative sets with different numbers of steps. In addition, some modules (such as M00611, the module representing photosynthesis), are made up of other modules, in which case they are only complete if their component modules are complete.
Hopefully this information will help you understand our estimation strategies in the next section.
KOfam annotations
For metabolism estimation to work properly, gene identifiers in the pool of annotations must match to the gene identifiers used in the pathway definitions. For KEGG MODULEs, we rely on annotations from the KEGG KOfam database, which is a set of HMM profiles for KEGG Orthologs (KOs). The program anvi-run-kegg-kofams can annotate your contigs-db with hits to the KEGG KOfam database. It adds these annotations under the source name ‘KOfam’.
Which of the annotations are considered for metabolism estimation depends on the input context. If you are working with isolate genomes (ie, not metagenome mode or bins), then all of the annotations under source ‘KOfam’ will be used. If you are working with bins in metagenomes, then for each bin, only the ‘KOfam’ annotations that are present in that bin will be in the annotation pool. Finally, for metagenome mode, since estimation is done for each contig separately, only the annotations present in each contig will be considered at a time.
How is the module completeness score calculated?
For demonstration purposes, let’s talk through the estimation of module completeness for one module, in one ‘sample’ (ie a genome, bin, or contig in a metagenome). Just keep in mind that the steps described below are followed for each module in each sample.
Step 1: Unrolling module definitions
As you saw above in the module examples, there can be multiple alternative KOs for a given step in a pathway. This means that there can be more than one way to have a ‘complete’ metabolic module. Therefore, to estimate completeness, we first have to identify all possible ‘paths’ through the module definition, where a ‘path’ is a set of KOs that could make the module complete (if they were all present in the annotation pool).
anvi-estimate-metabolism uses a recursive algorithm to “unroll” the module definition string into a list of all possible paths. First, the definition string is split into its component steps (which are separated by spaces). Each step is either an atomic step, a protein complex (KO components separated by ‘+’ or ‘-‘), or a compound step (multiple alternatives, separated by commas). Compound steps and protein complexes are recursively broken down until we have only atomic steps. An atomic step can be a single KO, a module number, a nonessential KO starting with ‘-‘, or ‘–’ (a string indicating that there is a reaction for which we do not have a KO). We use the atomic steps to build a list of alternative paths through the module definition. Protein complexes are split into their respective components using this strategy to find all possible alternative complexes, and then these complexes (with all their component KOs) are used to build the alternative paths.
Let’s see this in action, using the Threonine Biosynthesis module from above as an example. We first split the definition on spaces to get all component steps. Here we show each component step on its own line:
(K00928,K12524,K12525,K12526)
K00133
(K00003,K12524,K12525)
(K00872,K02204,K02203)
K01733
The first step is made up of 4 alternative KOs. We split on the commas to get these, and thus we have the starting KO for 4 possible alternative paths:
K00928 K12524 K12525 K12526
| | | |
The second step, K00133, is already an atomic step, so we can simply extend each of the paths with this KO:
K00928 K12524 K12525 K12526
| | | |
K00133 K00133 K00133 K00133
The third step is another compound step, but this time we can get 3 atomic steps out of it. That means that our 4 possible paths so far each gets 3 alternatives, bringing our total alternative path count up to 12:
K00928 K12524 K12525 K12526
| | | |
K00133 K00133 K00133 K00133
/ | \ / | \ / | \ / | \
K00003 K12524 K12525 K00003 K12524 K12525 K00003 K12524 K12525 K00003 K12524 K12525
Okay, hopefully you get the picture by now. The end result is a list of lists, like this:
[[K00928,K00133,K00003,K00872,K01733],
[K00928,K00133,K00003,K02204,K01733],
......
[K12526,K00133,K12525,K02203,K01733]]
in which every inner list is one of the alternative paths through the module definition - one of the possible ways to have a complete module.
By the way, here is one alternative path from the module M00011, just so you know what these look like with protein complexes:
[K00164+K00658+K00382,K01902+K01903,K00234+K00235+K00236+K00237,K01676,K00026]
Step 2: Marking steps complete
Once we have our list of alternative paths through the module, the next task is to compute the completeness of each path. Each alternative path is a list of atomic steps or protein complexes. We loop over every step in the path and use the annotation pool of KOs to decide whether the step is complete (1) or not (0). We have the following cases to handle:
-
A single KO - this is easy. If we have an annotation for this KO in our pool of ‘KOfam’ annotations, then the step is complete. (In our current implementation, we don’t pay attention to multiple annotations of the same KO - we only care if there is at least one.)
-
A protein complex - remember that these are multiple KOs connected with ‘+’ (if they are essential components) or ‘-‘ (if they are non-essential)? Well, for these steps, we compute a fractional completeness based on the number of essential components that are present in the annotation pool. We basically ignore the non-essential KOs. For example, the complex ‘K00174+K00175-K00177-K00176’ would be considered 50% complete (a score of 0.5) if only ‘K00174’ were present in the annotation pool.
-
Non-essential KOs - some KOs are marked as non-essential even when they are not part of a protein complex. They look like this: ‘-K12420’, with a minus sign in front of the KO identifier (that particular example comes from module M00778). These steps are ignored for the purposes of computing module completeness.
-
Steps without associated KOs - some reactions do not have a KO identifier, but instead there is the string ‘–’ serving as a placeholder in the module definition. Since we can’t annotate the genes required for these steps, we have no idea if they are complete or not, so we always consider them incomplete. Modules that have steps like this can therefore never have 100% completeness - it is sad, but what can we do? We warn the user about these instances so that they can check manually for any missing steps.
-
Modules - finally, some modules are defined by other modules. We can’t determine if these steps are complete until we’ve estimated completeness for every module, so we ignore these for now.
We add up the completeness of each essential step and divide by the number of essential steps to get the completeness score for a given path through the module.
Step 3: Module completeness
By this time, we have a completeness score (a fraction between 0 and 1) for every possible path through the module. To get the completeness score for the module overall, we simply take the maximum of all these completeness scores.
We can then check this number against the module completeness threshold (which is 0.75 by default). If the module completeness score is greater than or equal to the threshold, we mark the module as ‘complete’. This boolean value is meant only as a way to easily filter through the modules output, and you shouldn’t put too much stock in it because it covers up a lot of nuances, as you can tell from the details above :).
Step 4: Adjusting completeness
But don’t forget that there are some modules defined by other modules. These are usually what KEGG calls ‘Signature Modules’, which are collections of KOs that collectively encode some phenotype, rather than a typical pathway of chemical reactions. For these modules, we have to go back and adjust the completeness score after we know the completeness of its component modules. To do this, we basically re-do the previous two tasks to recompute the number of complete steps in each path and the overall completeness of the module. This time, when we reach a ‘Module’ atomic step (case 5), we take that module’s fractional completeness score to be the completeness of the step.
As an example, consider module M00618, the Acetogen signature module. Its definition is
M00377 M00579
Suppose module M00377 had a completeness score of 0.7 and module M00579 had a score of 0.4, based on the prior estimations. Then the completeness score of the [M00377,M00579] path would be (0.7+0.4)/2 = 0.55. Since this is the only possible path through the module, M00618 is 55% complete.
Summary
For those who prefer the less long-winded approach: module completeness in a given sample is calculated as the maximum fraction of essential KOs that are annotated in the sample, where the maximum is taken over all possible sets of KOs from the module definition.
Edit this file to update this information.
Additional Resources
Are you aware of resources that may help users better understand the utility of this program? Please feel free to edit this file on GitHub. If you are not sure how to do that, find the __resources__ tag in this file to see an example.