kegg-metabolism [artifact]
Table of Contents
![]()
A TXT-type anvi’o artifact. This artifact is typically generated, used, and/or exported by anvi’o (and not provided by the user)..
Back to the main page of anvi’o programs and artifacts.
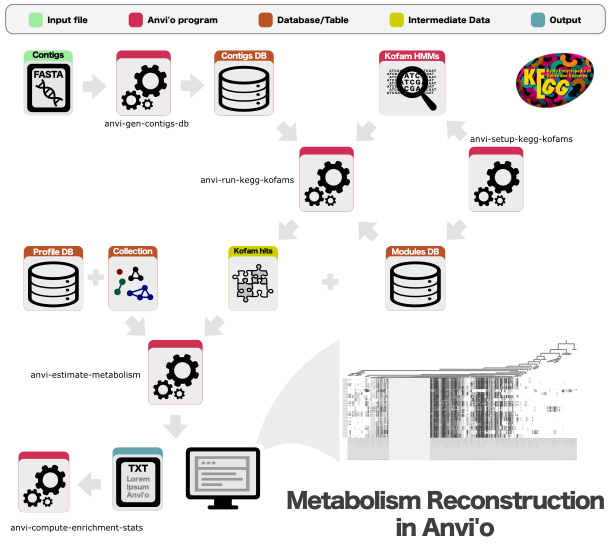
Provided by
Required or used by
anvi-compute-functional-enrichment
Description
Output text files produced by anvi-estimate-metabolism that describe the presence of metabolic pathways in a contigs-db.
Depending on the output options used when running anvi-estimate-metabolism, these files will have different formats.
How to get to this output
Long format output modes
The long format output modes produce tab-delimited files. Each line in the file (except for the header line) is indexed by an integer in the unique_id column. Different output “modes” will result in output files with different information.
‘Modules’ Mode
The ‘modules’ output file will have the suffix modules.txt. Each line in the file will represent information about a KEGG module in a given genome, metagenome, or bin. Here is one example, produced by running metabolism estimation on the Infant Gut dataset:
| unique_id | db_name | genome_name | kegg_module | module_name | module_class | module_category | module_subcategory | module_definition | kofam_hits_in_module | gene_caller_ids_in_module | module_completeness | module_is_complete |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | E_faecalis_6240 | Enterococcus_faecalis_6240 | M00001 | Glycolysis (Embden-Meyerhof pathway), glucose => pyruvate | Pathway modules | Carbohydrate metabolism | Central carbohydrate metabolism | ””“(K00844,K12407,K00845,K00886,K08074,K00918) (K01810,K06859,K13810,K15916) (K00850,K16370,K21071,K00918) (K01623,K01624,K11645,K16305,K16306) K01803 ((K00134,K00150) K00927,K11389) (K01834,K15633,K15634,K15635) K01689 (K00873,K12406)””” | K01834,K00134,K00873,K01689,K01624,K01803,K00927,K00850,K01810,K00845 | 2342,2646,1044,642,226,1041,348,1042,1043,225,600,1608 | 1.0 | True |
| 1 | E_faecalis_6240 | Enterococcus_faecalis_6240 | M00002 | Glycolysis, core module involving three-carbon compounds | Pathway modules | Carbohydrate metabolism | Central carbohydrate metabolism | ”"”K01803 ((K00134,K00150) K00927,K11389) (K01834,K15633,K15634,K15635) K01689 (K00873,K12406)””” | K01834,K00134,K00873,K01689,K01803,K00927 | 2342,2646,1044,642,226,1041,1042,1043 | 1.0 | True |
| (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) |
‘KOfam Hits in Modules’ Mode
The ‘kofam_hits_in_modules’ output file will have the suffix kofam_hits_in_modules.txt. Each line in the file will represent information about one KOfam hit in a given genome, metagenome, or bin - but ONLY KOs that are a part of at least one KEGG Module are included in this output. Hits are organized according to the KEGG module that they belong to, and more specifically the path through the KEGG module in which the KO appears.
What is a path through a KEGG module, you ask? Well. KEGG modules are metabolic pathways defined by a set of KOs. For example, here is the definition of module M00001, better known as “Glycolysis (Embden-Meyerhof pathway), glucose => pyruvate”:
(K00844,K12407,K00845,K00886,K08074,K00918) (K01810,K06859,K13810,K15916) (K00850,K16370,K21071,K00918) (K01623,K01624,K11645,K16305,K16306) K01803 ((K00134,K00150) K00927,K11389) (K01834,K15633,K15634,K15635) K01689 (K00873,K12406)
Spaces separate steps (reactions) in the metabolic pathway, and commas separate alternative KOs or alternative sub-pathways that can facilitate the same overall reaction. So a definition such as the one above can be “unrolled” into several different linear sequences of KOs, each of which we consider to be a possible “path” through the module. As an example, we can take the first option for every step in the Embden-Meyerhof pathway definition from above:
(K00844,K12407,K00845,K00886,K08074,K00918) (K01810,K06859,K13810,K15916) (K00850,K16370,K21071,K00918) (K01623,K01624,K11645,K16305,K16306) K01803 ((K00134,K00150) K00927,K11389) (K01834,K15633,K15634,K15635) K01689 (K00873,K12406)
to get the following path of KOs (which happens to be the path shown in the output example below):
K00844 K01810 K00850 K01623 K01803 K00134 K00927 K01834 K01689 K00873
For every KO in the path above that has a hit in the contigs-db, there will be a corresponding line in the ‘kofam_hits’ output file. The same will occur for every possible path in every single KEGG module, resulting in a lot of lines and extremely repetitive but nicely parseable information.
Without further ado, here is an example of this output mode (also from the Infant Gut dataset):
| unique_id | db_name | kofam_hit | gene_caller_id | contig | path_id | path | path_completeness | genome_name | kegg_module | module_completeness | module_is_complete |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | E_faecalis_6240 | K01834 | 2342 | Enterococcus_faecalis_6240_contig_00003_chromosome | 0 | K00844,K01810,K00850,K01623,K01803,K00134,K00927,K01834,K01689,K00873 | 0.8 | Enterococcus_faecalis_6240 | M00001 | 1.0 | True |
| 1 | E_faecalis_6240 | K01834 | 2646 | Enterococcus_faecalis_6240_contig_00003_chromosome | 0 | K00844,K01810,K00850,K01623,K01803,K00134,K00927,K01834,K01689,K00873 | 0.8 | Enterococcus_faecalis_6240 | M00001 | 1.0 | True |
| (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) |
‘KOfam Hits’ Mode
The kofam_hits output file will have the suffix kofam_hits.txt. Unlike the previous mode, this output will include ALL KOs, regardless of whether they belong to a KEGG Module or not. However, since only a subset of these KOs belong to modules, this output does not include module-related information like paths and module completeness.
Here is an example of this output mode (also from the Infant Gut dataset):
| unique_id | db_name | genome_name | ko | gene_caller_id | contig | modules_with_ko | definition |
|---|---|---|---|---|---|---|---|
| 0 | E_faecalis_6240 | Enterococcus_faecalis_6240 | K00845 | 1608 | Enterococcus_faecalis_6240_contig_00003_chromosome | M00001,M00549,M00892,M00909 | glucokinase [EC:2.7.1.2] |
| 1 | E_faecalis_6240 | Enterococcus_faecalis_6240 | K01810 | 600 | Enterococcus_faecalis_6240_contig_00003_chromosome | M00001,M00004,M00114,M00892,M00909 | glucose-6-phosphate isomerase [EC:5.3.1.9] |
| 2 | E_faecalis_6240 | Enterococcus_faecalis_6240 | K00850 | 225 | Enterococcus_faecalis_6240_contig_00003_chromosome | M00001,M00345 | 6-phosphofructokinase 1 [EC:2.7.1.11] |
| (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) |
Custom Mode (for module data)
The ‘modules_custom’ output mode will have user-defined content and the suffix modules_custom.txt (we currently only support output customization for modules data). See anvi-estimate-metabolism for an example command to work with this mode.
Matrix format output
Matrix format is an output option when anvi-estimate-metabolism is working with multiple contigs databases at once. The purpose of this output mode is to generate matrices of KEGG module statistics for easy visualization. Currently, the output of this mode includes a module completeness matrix, a matrix of binary module presence/absence values, and a matrix of KO counts. In these matrices, each row is a KEGG module or KO, and each column is an input contigs-db.
Here is an example of a module completeness matrix:
| module | bin_1 | bin_2 | bin_3 | bin_4 | bin_5 | bin_6 |
|---|---|---|---|---|---|---|
| M00001 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 |
| M00002 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| M00003 | 0.88 | 0.00 | 1.00 | 0.75 | 1.00 | 0.88 |
| M00004 | 0.88 | 0.00 | 0.88 | 0.88 | 0.88 | 0.00 |
| M00005 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| (…) | (…) | (…) | (…) | (…) | (…) | (…) |
While the above is the default matrix format, some users may want to include more annotation information in the matrices so that it is easier to know what is going on when looking at the matrix data manually. You can add this metadata to the matrices by using the --include-metadata flag when running anvi-estimate-metabolism, and the output will look something like the following:
| module | module_name | module_class | module_category | module_subcategory | bin_1 | bin_2 | bin_3 | bin_4 | bin_5 | bin_6 |
|---|---|---|---|---|---|---|---|---|---|---|
| M00001 | Glycolysis (Embden-Meyerhof pathway), glucose => pyruvate | Pathway modules | Carbohydrate metabolism | Central carbohydrate metabolism | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 |
| M00002 | Glycolysis, core module involving three-carbon compounds | Pathway modules | Carbohydrate metabolism | Central carbohydrate metabolism | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| M00003 | Gluconeogenesis, oxaloacetate => fructose-6P | Pathway modules | Carbohydrate metabolism | Central carbohydrate metabolism | 0.88 | 0.00 | 1.00 | 0.75 | 1.00 | 0.88 |
| (…) | (…) | (…) | (…) | (…) | (…) | (…) |
Edit this file to update this information.