kegg-metabolism
Table of Contents
![]()
A TXT-type anvi’o artifact. This artifact is typically generated, used, and/or exported by anvi’o (and not provided by the user)..
🔙 To the main page of anvi’o programs and artifacts.
Provided by
Required or used by
anvi-compute-metabolic-enrichment
Description
Output text files produced by anvi-estimate-metabolism that describe the presence of metabolic pathways in a contigs-db.
Depending on the output options used when running anvi-estimate-metabolism, these files will have different formats. This page describes and provides examples of the various output file types.
Please note that the examples below show only KEGG data, but user-defined metabolic pathways (user-metabolism) can also be included in this output!
How to get to this output
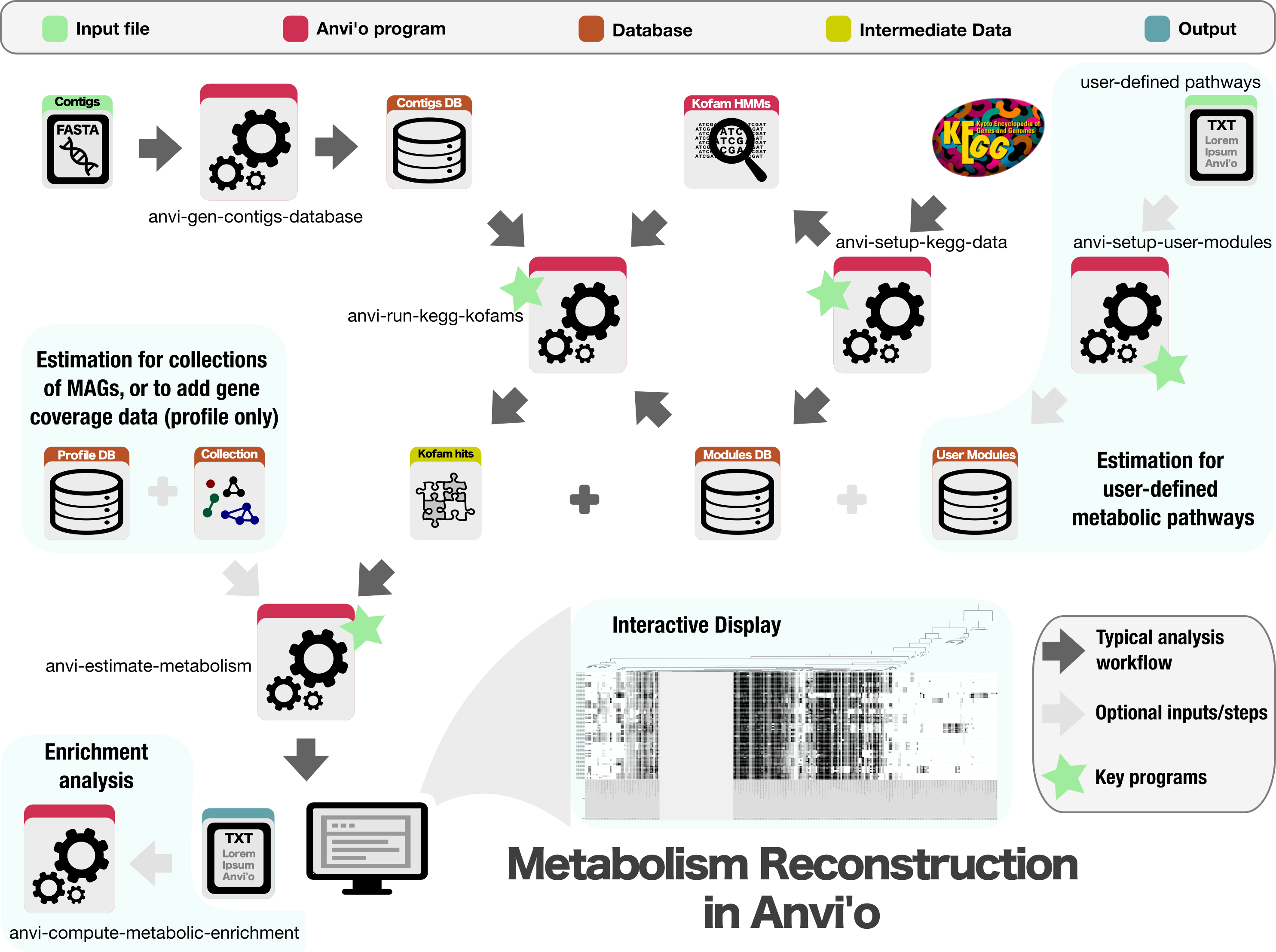
Long-format output modes
The long-format output option produces tab-delimited files. Different output “modes” will result in output files with different information. You can use the --list-available-modes parameter to see which modes are implemented in your version of anvi’o.
Some of these modes are customizable, such that you can select which columns of information to include in the output with the flag --custom-output-headers. Use the --list-available-output-headers parameter to see what kinds of information you can choose from.
‘Modules’ Mode
The ‘modules’ mode output file will have the suffix modules.txt. Each line in the file will represent information about a metabolic module in a given genome, bin, or contig of a metagenome assembly. Here is one example, produced by running metabolism estimation on the Enterococcus external genomes in the Infant Gut dataset:
| module | genome_name | db_name | module_name | module_class | module_category | module_subcategory | module_definition | stepwise_module_completeness | stepwise_module_is_complete | pathwise_module_completeness | pathwise_module_is_complete | proportion_unique_enzymes_present | enzymes_unique_to_module | unique_enzymes_hit_counts | enzyme_hits_in_module | gene_caller_ids_in_module | warnings |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | Glycolysis (Embden-Meyerhof pathway), glucose => pyruvate | Pathway modules | Carbohydrate metabolism | Central carbohydrate metabolism | “(K00844,K12407,K00845,K25026,K00886,K08074,K00918) (K01810,K06859,K13810,K15916) (K00850,K16370,K21071,K00918) (K01623,K01624,K11645,K16305,K16306) K01803 ((K00134,K00150) K00927,K11389) (K01834,K15633,K15634,K15635) K01689 (K00873,K12406)” | 1.0 | True | 1.0 | True | NA | No enzymes unique to module | NA | K00134,K00134,K00850,K00873,K00927,K01624,K01689,K01803,K01803,K01810,K01834,K01834,K25026 | 1044,642,225,226,1043,348,1041,1042,1043,600,2342,2646,1608 | K00850 is present in multiple modules: M00001/M00345,K00927 is present in multiple modules: M00001/M00002/M00003/M00308/M00552/M00165/M00166/M00611/M00612,K00873 is present in multiple modules: M00001/M00002,K01689 is present in multiple modules: M00001/M00002/M00003/M00346,K01624 is present in multiple modules: M00001/M00003/M00165/M00167/M00345/M00344/M00611/M00612,K01803 is present in multiple modules: M00001/M00002/M00003,K00134 is present in multiple modules: M00001/M00002/M00003/M00308/M00552/M00165/M00166/M00611/M00612,K01834 is present in multiple modules: M00001/M00002/M00003,K25026 is present in multiple modules: M00001/M00549/M00909,K01810 is present in multiple modules: M00001/M00004/M00892/M00909 |
| M00002 | Enterococcus_faecalis_6240 | E_faecalis_6240 | Glycolysis, core module involving three-carbon compounds | Pathway modules | Carbohydrate metabolism | Central carbohydrate metabolism | “K01803 ((K00134,K00150) K00927,K11389) (K01834,K15633,K15634,K15635) K01689 (K00873,K12406)” | 1.0 | True | 1.0 | True | NA | No enzymes unique to module | NA | K00134,K00134,K00873,K00927,K01689,K01803,K01803,K01834,K01834 | 1044,642,226,1043,1041,1042,1043,2342,2646 | K00927 is present in multiple modules: M00001/M00002/M00003/M00308/M00552/M00165/M00166/M00611/M00612,K00873 is present in multiple modules: M00001/M00002,K01689 is present in multiple modules: M00001/M00002/M00003/M00346,K01803 is present in multiple modules: M00001/M00002/M00003,K00134 is present in multiple modules: M00001/M00002/M00003/M00308/M00552/M00165/M00166/M00611/M00612,K01834 is present in multiple modules: M00001/M00002/M00003 |
| M00003 | Enterococcus_faecalis_6240 | E_faecalis_6240 | Gluconeogenesis, oxaloacetate => fructose-6P | Pathway modules | Carbohydrate metabolism | Central carbohydrate metabolism | “(K01596,K01610) K01689 (K01834,K15633,K15634,K15635) K00927 (K00134,K00150) K01803 ((K01623,K01624,K11645) (K03841,K02446,K11532,K01086,K04041),K01622)” | 0.8571428571428571 | True | 0.875 | True | NA | No enzymes unique to module | NA | K00134,K00134,K00927,K01624,K01689,K01803,K01803,K01834,K01834,K04041 | 1044,642,1043,348,1041,1042,1043,2342,2646,617 | K04041 is present in multiple modules: M00003/M00611/M00612,K00927 is present in multiple modules: M00001/M00002/M00003/M00308/M00552/M00165/M00166/M00611/M00612,K01689 is present in multiple modules: M00001/M00002/M00003/M00346,K01624 is present in multiple modules: M00001/M00003/M00165/M00167/M00345/M00344/M00611/M00612,K01803 is present in multiple modules: M00001/M00002/M00003,K00134 is present in multiple modules: M00001/M00002/M00003/M00308/M00552/M00165/M00166/M00611/M00612,K01834 is present in multiple modules: M00001/M00002/M00003 |
| (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) |
What are the data in each of these columns?
module: the module identifier for a metabolic pathway (from the KEGG MODULE database or from user-defined modules)genome_name/bin_name/contig_name: the identifier for the current sample, whether that is a genome, bin, or contig from a metagenome assemblydb_name: the name of the contigs database from which this data comes (only appears in output from multi-mode, in which multiple DBs are processed at once)module_name/module_class/module_category/module_subcategory/module_definition: metabolic pathway information, from the KEGG MODULE database or from user-defined metabolic modulesstepwise_module_completeness/pathwise_module_completeness: a fraction between 0 and 1 indicating the proportion of steps in the metabolic pathway that have an associated enzyme annotation. There are currently two strategies for defining the ‘steps’ in a metabolic pathway - ‘stepwise’ and ‘pathwise’. To learn how these numbers are calculated, see the anvi-estimate-metabolism help pagestepwise_module_is_complete/pathwise_module_is_complete: a Boolean value indicating whether the correspondingmodule_completenessscore is above a certain threshold or not (the default threshold is 0.75)proportion_unique_enzymes_present: some enzymes only belong to one metabolic pathway, which means that their presence is a better indicator for the presence of a metabolic capacity than other enzymes that are shared across multiple pathways. This column contains the fraction of these unique enzymes that are present in the sample. For instance, if the module has only 1 unique enzyme and it is present, you will see a 1 in this column. You can find out the denominator of this fraction (ie, the number of unique enzymes for this module) by either calculating the length of the list in theenzymes_unique_to_modulecolumn, or by requesting custom modules mode output with theunique_enzymes_context_stringheaderenzymes_unique_to_module: a comma-separated list of the enzymes that only belong to the current module (ie, are not shared across multiple metabolic pathways)unique_enzymes_hit_counts: a comma-separated list of how many times each unique enzyme appears in the sample, in the same order as theenzymes_unique_to_modulelistenzyme_hits_in_module: a comma-separated list of the enzyme annotations that were found in the current sample and contribute to this metabolic pathway (these will be enzymes from the metabolic pathway definition in themodule_definitioncolumn)gene_caller_ids_in_module: a comma-separated list of the genes with enzyme annotations that contribute to this pathway, in the same order as the annotations in theenzyme_hits_in_modulecolumnwarnings: miscellaneous caveats to consider when interpreting themodule_completenessscores. For example, a warning like “No KOfam profile for K00172” would indicate that we cannot annotate K00172 because we have no HMM profile for that gene family, which means that any metabolic pathway containing this KO can never be fully complete (even if a gene from that family does exist in your sequences). Seeing many warnings like “K01810 is present in multiple modules: M00001/M00004/M00892/M00909” indicates that the current module shares many enzymes with other metabolic pathways, meaning that it may appear to be complete only because its component enzymes are common. Extra caution should be taken when considering the completeness of modules with warnings
Module copy number values in the output
If you use the flag --add-copy-number when running anvi-estimate-metabolism, you will see three additional columns describing the estimated copy number of each module: pathwise_copy_number, stepwise_copy_number, and per_step_copy_numbers.
pathwise_copy_numberis the number of ‘complete’ copies of the path through the module with the highest (pathwise) completeness score, where ‘complete’ here means ‘greater than or equal to the module completeness threshold’. If there are multiple paths through the module with the highest pathwise completeness score, we take the maximum copy number of all of these paths.stepwise_copy_numberis the minimum number of times we see each top-level step in the module.per_step_copy_numbersis a comma-separated list of the copy number of each individual top-level step in the module. It is meant to be used for interpreting the stepwise copy number (which is simply the minimum of this list).
A discussion of how copy numbers are computed can be found here.
Coverage and detection values in the output
If you use the flag --add-coverage and provide a profile database, additional columns containing coverage and detection data will be added for each sample in the profile database. Here is a mock example of the additional columns you will see (for a generic sample called ‘SAMPLE_1’):
| SAMPLE_1_gene_coverages | SAMPLE_1_avg_coverage | SAMPLE_1_gene_detection | SAMPLE_1_avg_detection |
|---|---|---|---|
| 3.0,5.0,10.0,2.0 | 5.0 | 1.0,1.0,1.0,1.0 | 1.0 |
In this mock example, the module in this row has four gene calls in it. The SAMPLE_1_gene_coverages column lists the mean coverage of each of those genes in SAMPLE_1 (in the same order as the gene calls are listed in the gene_caller_ids_in_module column), and the SAMPLE_1_avg_coverage column holds the average of these values. As you probably expected, the detection columns are similarly defined, except that they contain detection values instead of coverage.
Pathway substrates, products, and intermediates
To add information about molecular compounds that are relevant to each metabolic pathway, you can customize the modules mode output. Here is an example command to do that:
anvi-estimate-metabolism -c contigs-db \ --output-modes modules_custom \ --custom-output-headers module,module_name,module_substrates,module_intermediates,module_products,pathwise_module_completeness,stepwise_module_completeness
The resulting output file will have a column for each item in the --custom-output-headers list, including one each for the substrates (input compounds), products (output compounds) and intermediates.
The ‘hits_in_modules’ output mode has been deprecated as of anvi’o v7.1-dev. If you have one of these output files and need information about it, you should look in the documentation pages for anvi’o v7. If you would like to obtain a similar output, the closest available is ‘module_paths’ mode.
‘Module Paths’ Mode
The ‘module_paths’ output file will have the suffix module_paths.txt. Each line in the file will represent information about one path through a metabolic module.
What is a path through a module, you ask? Well. There is a lengthier explanation of this here, but we will go through it briefly below.
Modules are metabolic pathways defined by a set of enzymes - for KEGG modules, these enzymes are KEGG orthologs, or KOs. For example, here is the definition of module M00001, better known as “Glycolysis (Embden-Meyerhof pathway), glucose => pyruvate”:
(K00844,K12407,K00845,K00886,K08074,K00918) (K01810,K06859,K13810,K15916) (K00850,K16370,K21071,K00918) (K01623,K01624,K11645,K16305,K16306) K01803 ((K00134,K00150) K00927,K11389) (K01834,K15633,K15634,K15635) K01689 (K00873,K12406)
Spaces separate steps (reactions) in the metabolic pathway, and commas separate alternative KOs or alternative sub-pathways that can facilitate the same overall reaction. So a definition such as the one above can be “unrolled” into several different linear sequences of KOs, each of which we consider to be a possible “path” through the module. As an example, we can take the first option for every step in the Embden-Meyerhof pathway definition from above:
(K00844,K12407,K00845,K00886,K08074,K00918) (K01810,K06859,K13810,K15916) (K00850,K16370,K21071,K00918) (K01623,K01624,K11645,K16305,K16306) K01803 ((K00134,K00150) K00927,K11389) (K01834,K15633,K15634,K15635) K01689 (K00873,K12406)
to get the following path of KOs (which happens to be the first path shown in the output example below):
K00844 K01810 K00850 K01623 K01803 K00134 K00927 K01834 K01689 K00873
In summary, a ‘path’ is one set of enzymes that can be used to catalyze all reactions in a given metabolic pathway, and there can be many possible paths (containing different sets of alternative enzymes) for a module. For every possible path through a module, there will be a corresponding line in the ‘module_paths’ output file.
The same principle applies to user-defined metabolic modules, except that the enzymes can be from a variety of different annotation sources (not just KOfam).
Without further ado, here is an example of this output mode (also from the Infant Gut dataset):
| module | genome_name | db_name | pathwise_module_completeness | pathwise_module_is_complete | path_id | path | path_completeness | annotated_enzymes_in_path |
|---|---|---|---|---|---|---|---|---|
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 0 | K00844,K01810,K00850,K01623,K01803,K00134,K00927,K01834,K01689,K00873 | 0.8 | [MISSING K00844],K01810,K00850,[MISSING K01623],K01803,K00134,K00927,K01834,K01689,K00873 |
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 1 | K12407,K01810,K00850,K01623,K01803,K00134,K00927,K01834,K01689,K00873 | 0.8 | [MISSING K12407],K01810,K00850,[MISSING K01623],K01803,K00134,K00927,K01834,K01689,K00873 |
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 2 | K00845,K01810,K00850,K01623,K01803,K00134,K00927,K01834,K01689,K00873 | 0.8 | [MISSING K00845],K01810,K00850,[MISSING K01623],K01803,K00134,K00927,K01834,K01689,K00873 |
| (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) |
Many of the columns in this data overlap with the ‘modules’ mode columns; you can find descriptions of those in the previous section. Below are the descriptions of new columns in this mode:
path_id: a unique identifier of the current path through the modulepath: the current path of enzymes through the module (described above)path_completeness: a fraction between 0 and 1 indicating the proportion of enzymes in the current path that are annotated. To learn how this number is calculated, see the anvi-estimate-metabolism help pageannotated_enzymes_in_path: a list of enzymes in the current path that were annotated in the current sample (in same order as the path). If an enzyme is missing annotations, that is indicated with the string[MISSING (enzyme)].
Note that in this output mode, pathwise_module_completeness and pathwise_module_is_complete are the pathwise completeness scores of the module overall, not of a particular path through the module. These values will be repeated for all lines describing the same module.
Path copy number values in the output
If you use the flag --add-copy-number, this output mode will gain an additional column, num_complete_copies_of_path, which describes the number of ‘complete’ copies of the current path through the module. To calculate this, we look at the number of annotations for each enzyme in the path and figure out how many times we can use different annotations to get a copy of the path with a completeness score that is greater than or equal to the completeness score threshold. For more details, check out the anvi-estimate-metabolism help page
‘Module Steps’ Mode
The ‘module_steps’ output file will have the suffix module_steps.txt. Each line in the file will represent information about one top-level step in a metabolic module. The top-level steps are the major steps that you get when you split the module definition on a space. Each “top-level” step is comprised of one or more enzymes that either work together or serve as alternatives to each other to catalyze (usually) one reaction in the metabolic pathway.
If we use module M00001 as an example again, we would get the following top-level steps for this module:
- (K00844,K12407,K00845,K00886,K08074,K00918)
- (K01810,K06859,K13810,K15916)
- (K00850,K16370,K21071,K00918)
- (K01623,K01624,K11645,K16305,K16306)
- K01803
- ((K00134,K00150) K00927,K11389)
- (K01834,K15633,K15634,K15635)
- K01689
- (K00873,K12406)
The first top-level step in this module is comprised of different glucokinase enzymes, all of which can catalyze the conversion from alpha-D-Glucose to alpha-D-Glucose 6-phosphate. The second top-level step is made up of alternative KOs for the glucose-6-phosphate isomerase enzyme, which converts alpha-D-Glucose 6-phosphate to beta-D-Fructose 6-phosphate. And so on.
Each top-level step in a metabolic module gets its own line in the ‘module_steps’ output file. Here is an example showing all 9 steps for module M00001 in one Enterococcus genome from the Infant Gut Dataset:
| module | genome_name | db_name | stepwise_module_completeness | stepwise_module_is_complete | step_id | step | step_completeness |
|---|---|---|---|---|---|---|---|
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 0 | (K00844,K12407,K00845,K25026,K00886,K08074,K00918) | 1 |
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 1 | (K01810,K06859,K13810,K15916) | 1 |
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 2 | (K00850,K16370,K21071,K00918) | 1 |
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 3 | (K01623,K01624,K11645,K16305,K16306) | 1 |
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 4 | K01803 | 1 |
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 5 | ((K00134,K00150) K00927,K11389) | 1 |
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 6 | (K01834,K15633,K15634,K15635) | 1 |
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 7 | K01689 | 1 |
| M00001 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1.0 | True | 8 | (K00873,K12406) | 1 |
| (…) | (…) | (…) | (…) | (…) | (…) | (…) | (…) |
As in the previous section, you should look at the ‘modules’ mode section for descriptions of any columns that are shared with that mode. Below are the descriptions of new columns in this mode:
step_id: a unique identifier of each top-level step in the modulestep: the definition of the top-level step, as extracted from the module definitionstep_completeness: an integer value of 1 in this column indicates that the step is complete, meaning that (at least) one of any alternative enzymes (or sets of enzymes) in this step has been annotated. A value of 0 indicates that the step is incomplete, meaning that there is no way for the step’s reaction to be catalyzed based on the set of enzyme annotations we are considering. This value is binary (so 0 and 1 are the only possible values for this column). To learn how this number is calculated, see the anvi-estimate-metabolism help page.
Note that in this output mode, stepwise_module_completeness and stepwise_module_is_complete are the stepwise completeness scores of the module overall, not of a particular step in the module. These values will be repeated for all lines describing the same module.
Step copy number values in the output
If you use the flag --add-copy-number, this output mode will gain an additional column, step_copy_number, which describes the number of copies of the current step. To calculate this value, we look at the number of annotations for each alternative enzyme in the step and figure out how many different versions of the step we can make by combining different annotations. For more details, check out the anvi-estimate-metabolism help page.
Enzyme ‘Hits’ Mode
The ‘hits’ output file will have the suffix hits.txt. Unlike the previous mode, this output will include ALL enzyme hits (from all annotation sources used for metabolism estimation), regardless of whether the enzyme belongs to a metabolic module or not. Since only a subset of these enzymes belong to modules, this output does not include module-related information like paths and module completeness.
Here is an example of this output mode (also from the Infant Gut dataset):
| enzyme | genome_name | db_name | gene_caller_id | contig | modules_with_enzyme | enzyme_definition |
|---|---|---|---|---|---|---|
| K25026 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 1608 | Enterococcus_faecalis_6240_contig_00003_chromosome | M00001,M00549,M00909 | glucokinase [EC:2.7.1.2] |
| K01810 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 600 | Enterococcus_faecalis_6240_contig_00003_chromosome | M00001,M00004,M00892,M00909 | glucose-6-phosphate isomerase [EC:5.3.1.9] |
| K00850 | Enterococcus_faecalis_6240 | E_faecalis_6240 | 225 | Enterococcus_faecalis_6240_contig_00003_chromosome | M00001,M00345 | 6-phosphofructokinase 1 [EC:2.7.1.11] |
| (…) | (…) | (…) | (…) | (…) | (…) | (…) |
Here are the descriptions of any new columns not yet discussed in the previous sections:
enzyme: an enzyme that was annotated in the contigs databasemodules_with_enzyme: the modules (if any) that this enzyme belongs toenzyme_definition: the function of this enzyme (often includes the enzyme name and EC number)
Coverage and detection values in the output
If you use the flag --add-coverage and provide a profile database, you will get one additional column per sample for coverage (containing the coverage value of the enzyme annotation in the sample) and one additional column per sample for detection (containing the detection value of the enzyme annotation in the sample). Here is a mock example:
| SAMPLE_1_coverage | SAMPLE_1_detection |
|---|---|
| 3.0 | 1.0 |
Since each row is a single gene in this output mode, these columns will contain the coverage/detection values for that gene only.
Custom Mode (for module data)
The ‘modules_custom’ output mode will have user-defined content and the suffix modules_custom.txt (we currently only support output customization for modules data). See anvi-estimate-metabolism for an example command to work with this mode. The output file will look similar to the ‘modules’ mode output, but with a different (sub)set of columns. You can use the flag --list-available-output-headers to see all of the possible columns you can choose from - this list will change depending on what input type you have and whether you use the --add-copy-number or --add-coverage flags (one caveat: using these flags with ‘Multi Mode’ input does not show you all possible output headers, so it is best to build your custom header list by looking at the possible headers for one sample).
Matrix format output
Matrix format is an output option when anvi-estimate-metabolism is working with multiple contigs databases at once (otherwise known as ‘Multi Mode’). The purpose of this output type is to generate matrices of module statistics for easy visualization and clustering. Currently, the matrix-formatted output includes:
- matrices of module completeness scores, one for pathwise completeness and one for stepwise completeness
- matrices of binary module presence/absence values, one for pathwise completeness and one for stepwise completeness
- matrix of binary top-level step completeness values
- matrix of enzyme annotation counts
If you use the --add-copy-number flag, you will get three additional matrix files:
- matrices of module copy number, one for pathwise copy number and one for stepwise copy number
- matrix of top-level step copy number
In these tab-delimited matrix files, each row is a module, top-level step, or enzyme, and each column is an input sample.
Here is an example of a module pathwise completeness matrix, for bins in a metagenome:
| module | bin_1 | bin_2 | bin_3 | bin_4 | bin_5 | bin_6 |
|---|---|---|---|---|---|---|
| M00001 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 |
| M00002 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| M00003 | 0.88 | 0.00 | 1.00 | 0.75 | 1.00 | 0.88 |
| M00004 | 0.88 | 0.00 | 0.88 | 0.88 | 0.88 | 0.00 |
| M00005 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| (…) | (…) | (…) | (…) | (…) | (…) | (…) |
Each cell of the matrix is the pathwise completeness score for the corresponding module in the corresponding sample (which is, in this case, a bin).
While the above is the default matrix format, some users may want to include more annotation information in the matrices so that it is easier to know what is going on when looking at the matrix data manually. You can add this metadata to the matrices by using the --include-metadata flag when running anvi-estimate-metabolism, and the output will look something like the following:
| module | module_name | module_class | module_category | module_subcategory | bin_1 | bin_2 | bin_3 | bin_4 | bin_5 | bin_6 |
|---|---|---|---|---|---|---|---|---|---|---|
| M00001 | Glycolysis (Embden-Meyerhof pathway), glucose => pyruvate | Pathway modules | Carbohydrate metabolism | Central carbohydrate metabolism | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 |
| M00002 | Glycolysis, core module involving three-carbon compounds | Pathway modules | Carbohydrate metabolism | Central carbohydrate metabolism | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| M00003 | Gluconeogenesis, oxaloacetate => fructose-6P | Pathway modules | Carbohydrate metabolism | Central carbohydrate metabolism | 0.88 | 0.00 | 1.00 | 0.75 | 1.00 | 0.88 |
| (…) | (…) | (…) | (…) | (…) | (…) | (…) |
The module/step completeness matrix files will have the suffix completeness-MATRIX.txt.
Module presence/absence matrix files will have the suffix presence-MATRIX.txt. In these files, each cell of the matrix will have either a 1.0 or a 0.0. A 1.0 indicates that the module has a completeness score above the module completeness threshold in that sample, while a 0.0 indicates that the module’s completeness score is not above the threshold.
Enzyme hit matrix files will have the suffix enzyme_hits-MATRIX.txt. Each row of the matrix will be an enzyme, and each column will be an input sample. Cells in this matrix will contain an integer value, representing the number of times the enzyme was annotated in that sample. (Note: you will also add metadata to this matrix type when you use the --include-metadata flag).
Copy number matrices will have the suffix copy_number-MATRIX.txt. In these files, each cell of the matrix will be an integer representing the number of copies of a module or top-level step in a given sample.
Edit this file to update this information.