profile-db
Table of Contents
![]()
A DB-type anvi’o artifact. This artifact is typically generated, used, and/or exported by anvi’o (and not provided by the user)..
🔙 To the main page of anvi’o programs and artifacts.
Provided by
Required or used by
anvi-cluster-contigs anvi-db-info anvi-delete-collection anvi-delete-misc-data anvi-delete-state anvi-display-metabolism anvi-display-structure anvi-estimate-genome-completeness anvi-estimate-metabolism anvi-estimate-scg-taxonomy anvi-estimate-trna-taxonomy anvi-export-collection anvi-export-gene-coverage-and-detection anvi-export-items-order anvi-export-misc-data anvi-export-splits-and-coverages anvi-export-state anvi-gen-fixation-index-matrix anvi-gen-gene-consensus-sequences anvi-gen-gene-level-stats-databases anvi-gen-variability-profile anvi-get-aa-counts anvi-get-codon-frequencies anvi-get-codon-usage-bias anvi-get-sequences-for-hmm-hits anvi-get-short-reads-from-bam anvi-get-split-coverages anvi-import-collection anvi-import-items-order anvi-import-misc-data anvi-import-state anvi-inspect anvi-interactive anvi-merge-bins anvi-migrate anvi-refine anvi-rename-bins anvi-search-sequence-motifs anvi-show-collections-and-bins anvi-show-misc-data anvi-split anvi-summarize anvi-update-db-description anvi-script-add-default-collection anvi-script-gen-distribution-of-genes-in-a-bin anvi-script-gen-genomes-file anvi-script-permute-trnaseq-seeds
Description
An anvi’o database that contains key information about the mapping of short reads from multiple samples to your contigs.
You can think of this as a extension of a contigs-db that contains information about how your contigs align with each of your samples. The vast majority of programs that use a profile database will also ask for the contigs database associated with it.
A profile database contains information about how short reads map to the contigs in a contigs-db. Specifically, for each sample, a profile database contains
- the coverage and abundance per nucleotide position for each contig
- variants of various kinds (single-nucleotide, single-codon, and single-amino acid)
- structural variants (ex. insertions and deletions) These terms are explained on the anvi’o vocabulary page.
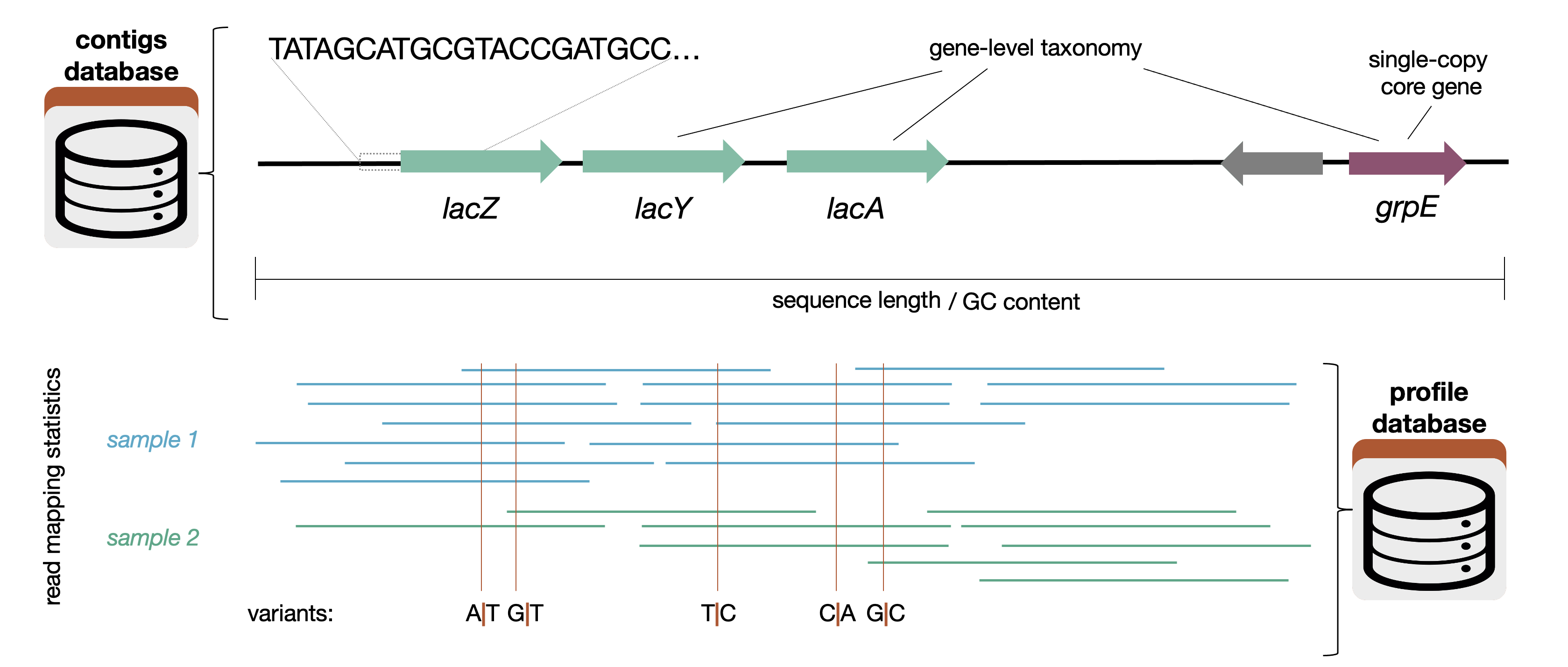
This information is necessary to run anvi’o programs like anvi-cluster-contigs, anvi-estimate-metabolism, and anvi-gen-gene-level-stats-databases. You can also interact with a profile database using programs like anvi-interactive.
Technically, “profile-db” refers to a profile database that contains the data from several samples – in other words, the result of running anvi-merge on several single-profile-db. However, since a single-profile-db has a lot of the functionality of a profile-db, it might be easier to think of a profile database as a header referring to both single-profile-dbs and profile-dbs (which can also be called a merged-profile-dbs). For simplicity’s sake, since most users are dealing with multiple samples, the name was shortened to just profile-db. The following are a list of differences in functionality between a single profile database and a merged profile database:
- You can run anvi-cluster-contigs or anvi-mcg-classifier on only a merged profile database (or profile-db), since they look at the allignment data in many samples
- You cannot run anvi-merge or anvi-import-taxonomy-for-layers on a merged profile database, only on a single-profile-db.
How to make a profile database
If you have multiple samples
- Prepare your contigs-db
- Run anvi-profile with an appropriate bam-file. The output of this will give you a single-profile-db. You will need to do this for each of your samples, which have been converted into a bam-file with your short reads.
- Run anvi-merge on your contigs-db (from step 1) and your single-profile-dbs (from step 2). The output of this is a profile-db.
If you have a single sample
- Prepare your contigs-db
- Run anvi-profile with an appropriate bam-file. The output of this will give you a single-profile-db. You can see that page for more information, but essentially you can use a single-profile-db instead of a profile database to run most anvi’o functions.
Variants
Profile databases, like contigs-dbs, are allowed to have different variants, though the only currently implemented variant, the trnaseq-profile-db, is for tRNA transcripts from tRNA-seq experiments. The default variant stored for “standard” profile databases is unknown. Variants should indicate that substantially different information is stored in the database. For instance, single codon variability is applicable to protein-coding genes but not tRNA transcripts, so SCV data is not recorded for the trnaseq variant. The $(trnaseq-workflow)s generates trnaseq-profile-dbs using a very different approach to anvi-profile.
Edit this file to update this information.